In one of his best loved poems, William Blake invited his readers to “see a world in a grain of sand.” His words are as inspiring as ever, but it must be said that his imagery is a little out of date. Today, we can see multiple universes—or “omes”—in a single cell. Visions that are at once more detailed and more expansive are opening before us because we now possess technologies that allow us to survey genomes, transcriptomes, proteomes, and other “omes.” What’s more, these technologies are being incorporated into multimodal omics platforms, giving us the ability to see relationships and patterns that would have remained unseen, had we been confined to single-ome views.
Single-cell multimodal omics is no poet’s fancy. It is being developed by several companies, and it is being embraced by the scientific community. For example, in 2019, it was designated Method of the Year by Nature Methods.
Multimodal omics at the single-cell level is “transformative,” declares Anjali Pradhan, vice president of product management at Mission Bio. She emphasizes that the technology opens new dimensions of biological understanding and has implications in personalized medicine. Single-cell multiomics, she points out, has already allowed researchers to make new clinical discoveries about disease characterization and behavior that will drive patient treatment and therapeutic development approaches.
Omics technologies are being multiplexed in various ways—as various as the companies that are in this fast-growing field. Some of the companies contributed their insights to this article. Despite pursuing different approaches, they have at least one thing in common: a desire to share their passion for endless discovery. Were the companies to wax poetic, they might, like Blake, invite you to “hold infinity in the palm of your hand, and eternity in an hour.”
Spanning genotype and phenotype
Although Mission Bio changed its CEO last year, the company’s focus has stayed the same. The decade-old single-cell technology company has maintained a focus on multimodal omics from the beginning. Mission Bio’s two-step droplet microfluidics technology, offered through the Tapestri platform, performs simultaneous single-cell DNA and protein analysis (to uncover both the genotype and phenotype in the same cell, at the same time.) The company is currently working on expanding the capabilities of the Tapestri to include RNA.

In April of last year, the company announced the appointment of Yan Zhang, PhD, to the role of CEO, taking over for co-founder Charlie Silver. Zhang spent the previous decade at Thermo Fisher Scientific, growing genetic analysis businesses. And she intends to do the same for multimodal omics while heading Mission Bio.
As Zhang wrote in an essay (“Why Multi-Omics Is the Future of Biological Analysis”) that was published October 22 in Forbes, the first area that could benefit from multiomics techniques is cancer. The advances could come from analyzing genotype-to-phenotype changes, understanding mechanisms of drug resistance, or increasing the general understanding of cancer-like diseases.
The technology is already moving into translational and clinical research. For example, the technology was used by Memorial Sloan Kettering researchers to study evolution of resistance against sotorasib, a KRAS inhibitor drug for lung cancer. The researchers attributed the evolution of resistance to secondary mutations within individual cancer cells (Nature 2021; 599: 679–683).
In addition, a team from St. Jude Children’s Research Hospital used the Tapestri to explore somatic genetic rescue in pediatric patients with myelodysplastic syndromes, disorders in which blood cells do not mature. In a recent article (Nat. Med. 2021; 27: 1806–1817), the researchers identified cells in pediatric patients carrying germline SAMD9/9L mutations that can self-correct without the need for a bone marrow transplant.
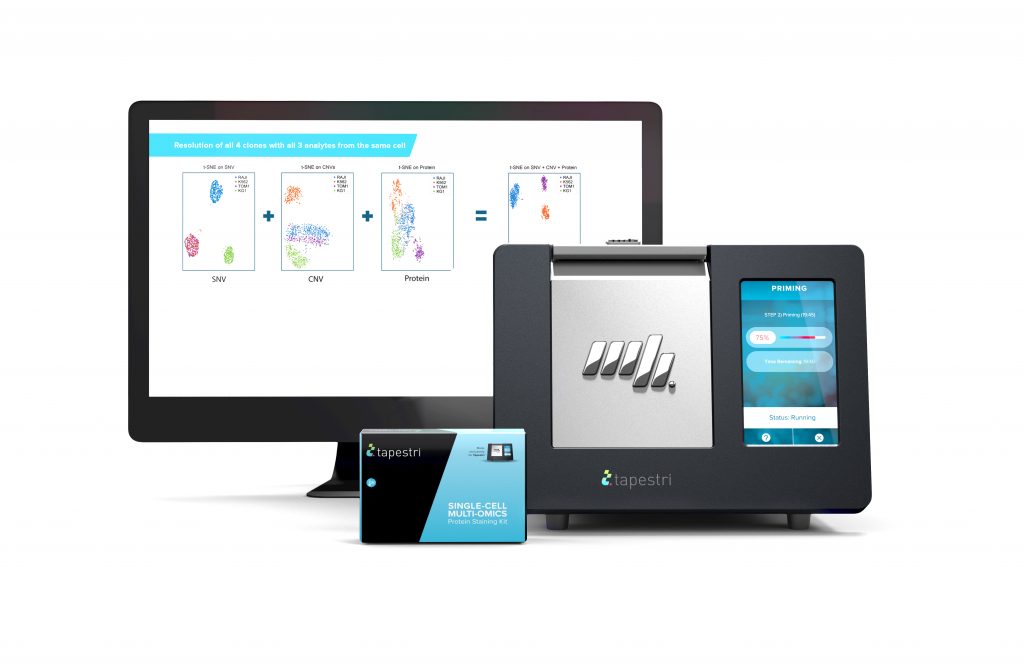
Another area where high-throughput, single-cell multiomic characterization could be useful is in advancing cell and gene therapies. The Tapestri could simplify the characterization of gene edited cells, measuring multiple attributes simultaneously in the same cells, before the cells head into the clinic.
Spanning the transcriptome and the proteome
After IsoPlexis launched its proteomics platform in 2018, the company saw that the most common request it received was for the ability to sequence the cells detected on the platform. Now, IsoPlexis has developed a way to do that. The company’s new Duomic technology adds RNA sequencing to the proteomics platform, measuring functional protein and gene expression levels from the same single cell. The mRNA capture, sample prep, and proteomic capture are performed on IsoPlexis’s chips. Then, the mRNA is examined downstream on a sequencer.
Sean Mackay, CEO and co-founder of IsoPlexis, tells GEN why the RNA addition is important. Getting to the single-cell level of understanding of which cells are most proteomically active is important for a variety of clinical applications, he notes. Accessing the genetic code for those proteomically driven cells, he adds, “can teach us how to edit out pathways and how to replicate cells using different types of genetic modifications like [those in] CAR T cells.”
Data from the Duomic was first presented at the Advances in Genome Biology and Technology (AGBT) precision health conference held in September 2021. (Duomic-driven data generation will be available as a service in the second half of this year.) Rui Zheng, PhD, IsoPlexis’s senior vice president of innovation, and Patrick Paczkowski, PhD, the company’s senior vice president of software, used the Duomic to simultaneously measure the protein and gene expression levels of 55 individual melanoma tumor cells.
The goal of the Duomic, they said, is to be able to better understand the gene networks that are regulating some of the most important and most problematic phosphoproteomically driven cells. They assert that the platform allows researchers to dissect these interactions and could lead to the tuning of problematic cells. In the presentation, which included analysis by Virtualitics’ AI-driven 3D data visualization software platform, the Duomic identified cell populations based on functional heterogeneity in addition to gene expression profiles driving the proteomics.
Where will the Duomic be most useful? Mackay thinks immunology and immune profiling are at the top of the list. More specifically, he thinks the Duomic will lead to reengineering of CAR T cells to optimize each cell, to understanding the complementary gene and T-cell receptor profiles of potent proteomic immune cells, and to being able to apply profiles of various types of immune cells.
Mackay sees a day when any researcher can access multiomics data routinely. Although IsoPlexis intends to add epigenomic and metabolomic capabilities to its offerings, the company is staying focused for the moment on the three core omics disciplines—genomics, transcriptomics, and proteomics.
One of the first companies to develop technology to simultaneously detect mRNA and proteins is BD Biosciences. The company’s Rhapsody Single-Cell Analysis System uses microwell-based technology to capture and barcode hundreds to thousands of single cells for analysis of genomic and proteomic information. The company notes that its well system, which relies on gravity-based settling of cells, allows for capture of diverse cell types including fragile cells such as neutrophils. After cells are sorted, beads are loaded into the wells. Upon cell lysis, the targets are captured by the beads, which are used to generate next-generation sequencing (NGS) libraries.
Spanning the genome and the proteome
SomaLogic is neither a single-cell company nor a multimodal omics company. It is firmly rooted in the proteomics space. For more than 20 years, the company has been refining its aptamer-based technology for measuring proteins. Most recently, SomaLogic has been arranging for its platform to interoperate with other platforms and play its part in the multimodal omics future.
In January 2022, SomaLogic announced that it was partnering with Illumina to combine the companies’ proteomics and genomics platforms. The goal? To develop co-exclusive, co-branded, NGS-based proteomics products. Illumina will develop and sell a kit, based on SomaLogic’s technology, for people who want to measure proteins using NGS.
The SomaScan platform uses SOMAmer reagents (Slow Off-Rate Modified Aptamers) to identify circulating proteins from a sample of plasma, serum, or urine. The reagents, synthesized with a fluorophore, photocleavable linker, and biotin, are bound to streptavidin beads. Proteins are bound and tagged with biotin. Exposure to ultraviolet light breaks the linker, releasing nonspecific complexes. The proteins can then be measured using a fluorescent array.

SomaScan technology has been described as an “unbiased and highly multiplexed search engine [that] will enable the discovery of novel biomarkers in a manner that is unencumbered by our incomplete knowledge of biology” (PLoS One 2010; 5(12):e15004). It is capable of measuring and identifying 7,000 human proteins—4,000 more proteins than antibody-based methods. SomaLogic estimates that it will be able to increase that number to 10,000—half of the human proteome—in a few years.
Roy Smythe, MD, Somalogic’s CEO, tells GEN, “The more proteins you can identify and measure, the more biology you can discern and the more targets you can develop.”
Spanning the genome and the epigenome
Bringing genomics on board makes sense for a proteomics company. But a genomics company like 10x Genomics must jump onto a different car of the multimodal omics train.
The first single-cell RNA-sequencing (scRNA-seq) study was published over a decade ago. Since that time, the technology at 10x Genomics has revolutionized single-cell genomics, with the company’s Chromium platform becoming a mainstay in molecular biology laboratories around the world.
The company has offered its RNA-seq kit separately from an ATAC-seq kit—for epigenomic analysis—for years. Researchers interested in analyzing both sets of data would require computational prediction, and inferred linkage, to match a single cell’s gene expression profile with its epigenome.
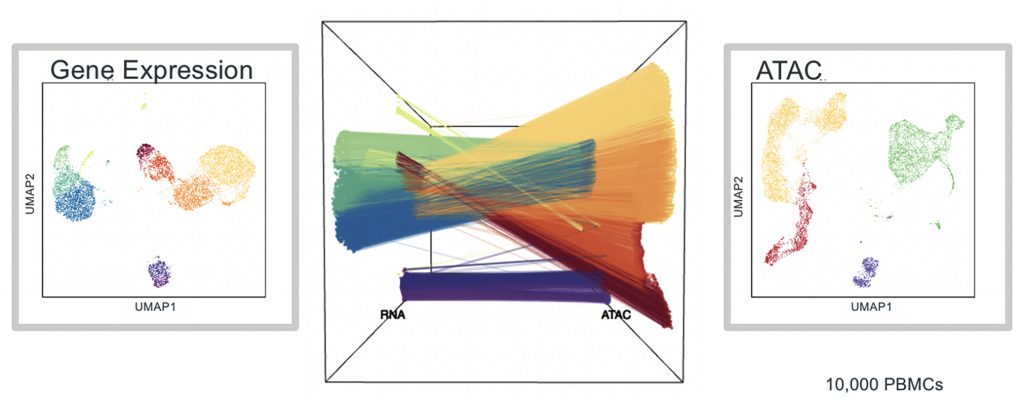
Wanting to help researchers overcome that hurdle, the company developed a kit capable of simultaneously profiling the epigenome and transcriptome from the same single cell. The Chromium Single Cell Multiome ATAC + Gene Expression kit was first previewed at the 2020 AGBT meeting in Marco Island, FL, and it was released later that year. Combining epigenome profiling and transcriptome profiling allows researchers to uncover new roles for regulatory elements in cell types and could have broad appeal to researchers in varied fields, including oncology, immunology, neuroscience, and stem cell and developmental biology.
Higher dimensions
There are many omes. They would be challenging to study even if they existed in parallel. As it is, they are intertwined. To see how—to explore higher dimensions—we can take advantage of multimodal omics. The technology is already showing us that explorations of higher dimensions can lead to down-to-earth gains.