The most exclusive club in omics has admitted only genomics and transcriptomics, the beneficiaries of single-molecule sequencing technologies that are so powerful they enable highly refined single-cell and spatial analyses. In contrast, proteomics has been underprivileged with respect to sequencing. But proteomics is about to enjoy a change in fortune. Multiple companies are developing single-molecule protein sequencing technology. As this technology enters the market, it is bound to give proteomics a higher profile.

Erisyon
“Genomics is so much easier than proteomics,” says Talli Somekh, co-founder and CEO of Erisyon. He explains that proteomics suffers from sheer complexity. Whereas genomics involves just four nucleotides, proteomics has to deal with 20 amino acids—and an untold number of post-translational modifications. And that’s not the only reason for the complexity. DNA and RNA are readily copied, whereas proteins lack such amplification. Also, the 20,000 to 25,000 genes in humans give rise to a hundred thousand transcripts, which in turn gives rise to millions of proteins. In addition, nucleic acids are easily extracted from solution, but proteins are more challenging because they have highly variable properties such as charge and pH. The biggest challenge, Somekh asserts, is dynamic range. In a single cell, the dynamic range of RNA is about 104; it’s about 109 for proteins.

Quantum-Si
Many companies have tried—and failed—to overcome these complexities. The result? A “graveyard of startups,” according to one investor. But new aspirants are undeterred. Grace Johnston, PhD, chief commercial officer at Quantum-Si, told GEN that proteomics can add to what genomics has taught us about disease. That is, information about disease that is beyond what genomics can reveal may need to be found in proteomics. She also suggests that proteomic information may be even more impactful than genomic information.
Mass spectrometry is here to stay
Mass spectrometry (MS) has been around for decades, and it is the workhorse of proteomics. But it has its limits. Areas of difficulty include post-translational modification (PTM) identification, working with very low sample concentrations, and performing single-cell proteomics. Other limits are in the detection of neoantigens on cancer cells, and in antibody sequencing. Lastly, MS is not cheap or easy to use.
The motivation behind protein sequencing is not to replace MS, but to complement it and allow researchers to dive deeper into proteomics. “We explicitly do not consider ourselves a competitor to MS or affinity assays,” Somekh remarks. He adds that MS and affinity assays are amazing technologies that are continually improving. “Our interest,” he emphasizes, “is to go where the limits block those technologies.”
Spin out
An erisyon, in ancient Greek, is a branch of olive or laurel decorated with figs, candies, tiny bottles of olive oil, wine, or honey. It represents an abundant future. At the eponymous protein sequencing startup, it is hoped peptide backbones decorated with fluorophores will make for a bright future.
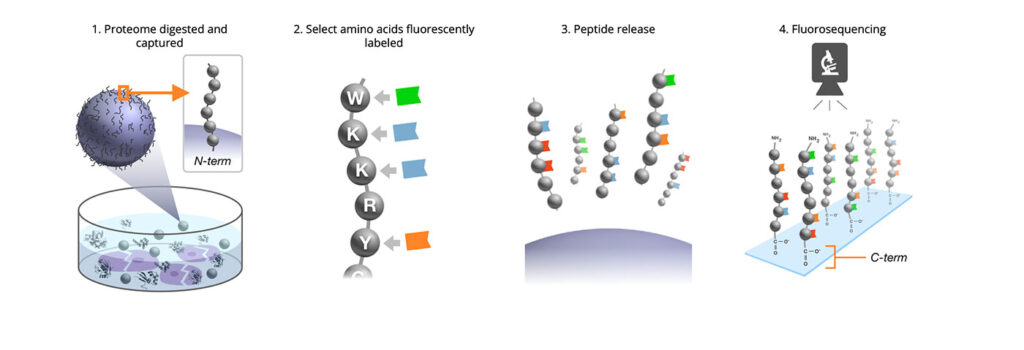
Erisyon asserts that it is commercializing the first single-molecule protein sequencer. The sequencer is based on the graduate work of Jag Swaminathan, PhD, the company’s chief technology officer. Swaminathan was in the laboratory of proteomics scientist Edward Marcotte, PhD, professor at UT Austin. Marcotte has been running an MS laboratory for 20 years, and he has—in Somekh’s telling—grown increasingly jealous of the fidelity and sensitivity that has been achieved by genomics researchers. Ultimately, Somekh, Swaminathan, and Marcotte joined together with Eric Anslyn, PhD, professor of chemistry at UT Austin, and Angela Bardo, PhD, a research scientist at the university, to found Erisyon in 2018.
The company’s technology determines how four amino acids contribute to a protein’s content. This information, the company’s founders say, suffices to establish a protein’s identity. (The founders also point to a proof-of-concept study they published in 2014.) They refer to the technology as the Peptide Wheel of Fortune: if you know the dictionary, the category that you’re looking for, and the positional information of a few letters, you know enough to solve the puzzle. The company maintains that proteomics scientists want to know more about the identity of proteins, and their quantity, than the full de novo sequence.
In 2018, the scientists who founded Erisyon described their technology—single-molecule fluorosequencing—in Nature Biotechnology. They emphasized that their technology is distinctive because it employs a process that begins with the covalent labeling of the amino acid side chains. (Many other companies use N-terminal affinity assays to determine the N-terminal amino acid.)
Erisyon adds fluorophores to the side chains prior to sequencing. After a sample of different proteins is digested into peptides, each of four amino acids is labeled with a fluorophore. Then, the peptides are immobilized by the C terminal to a glass flow cell. The next step is imaging by single-molecule microscopy (total internal reflective fluorescent microscopy). This is followed by classical Edman chemistry, which has been around since the 1950s. The peptide is exposed to a chemistry that removes an amino acid from the N-terminal end. Focusing on what is left after the fact, it is determined whether a step drop has occurred in any of the channels, and which amino acids were removed.
Like the other companies in this space, Erisyon aims to be a tool provider. Moreover, Erisyon plans to bring its technology to the market relatively soon. The company is working with a number of collaborators, and it has generated data that will likely be published. (The company has a history of vetting its work through the scientific literature.)
The language of protein
Like many people in 2020, Joshua Yang, and Daniel Estandian, PhD, met over Zoom. They were participating in Nucleate, which resembles an online speed dating program and pairs business students with scientists. After only a handful of meetings, the two decided to work together. They founded Glyphic Biotechnologies in March 2021, fundraised a $6 million seed round, and then met each other in person—eight months later.
The company is based on the technology developed by Estandian while he was a graduate student in the laboratory of Ed Boyden, PhD, a professor at the McGovern Institute at MIT and an HHMI investigator. Glyphic plans to launch early access for the technology next year. Initially, the technology will be service based. The long-term goal is to release an instrument at the end of 2025.

What sets Glyphic apart? “[It] will be the only one in the market that can actually do true de novo protein sequencing,” Yang says. “What I mean by that is sequencing [that accounts for all] 20 amino acids as well as post-translational modifications.” He says that this may be particularly useful for unknown proteins and proteins with variable regions or mutations.
Glyphic’s approach is similar to that of other companies in this space, in that Glyphic uses N-terminal binders. Sequencing all 20 amino acids brings a unique complication. It has not been possible to develop 20 specific N-terminal binders against 20 amino acids because of interfering amino acids (20 amino acids at the second position and another 20 amino acids in the third position) creating 400 possible local environments around any amino acid. Ordinarily, this is not a tractable problem, Yang explains.
Glyphic overcomes this problem by tethering the last amino acid onto a surface, attaching the peptide at the N- and C-termini. The N-terminus is a DNA-
mediated, reversible attachment that involves the ClickP molecule. The last amino acid is cleaved off and remains stuck on the surface by itself. Binders that can bind specifically, and with high affinity, are flowed over the lone amino acid. The binders are antibodies that can be conjugated to fluorophores or to barcodes—both of which the company is testing. The process is then continued sequentially.
This technology, which is published in Estandian’s doctoral thesis (but not a peer-reviewed publication), can also detect post-translational modifications through the development of new binders against each of the different modifications that people are interested in.
With 20 people working in New York City, Yang knows that it’s early days for Glyphic and the field. But he looks forward to a future when everyone will have access to protein sequencing instruments, much as they currently have access to next-generation sequencing instruments.
First out of the gate
If this field were in a race, Quantum-Si would be recognized as having been the first out of the gate. The company developed the first commercially available platform and started shipping its instrument, the Platinum, last January. The 200-person strong company, based in Guildford, CT, was founded in 2013 by Jonathan Rothberg, PhD, the founder of 454 Life Sciences and a pioneer of semiconductor-based sequencing. Quantum-Si’s protein sequencing is also semiconductor chip-based and uses kinetics to detect amino acids. Instead of one recognizer for each amino acid, Quantum-Si’s instrument has five recognizers, and they allow up to 15 amino acids to be detected. The recognizers are generated using directed evolution from the ClpS proteins that naturally bind to N-terminal amino acids.
Johnston says that her experiences running Western blots during her graduate work in cell biology served as motivation to join a company with a focus on making protein sequencing accessible to everyone. Using kinetics, Johnston insists, is the “best horse in the race.”
A protein is broken into peptides that bind to the bottom of the well (a single peptide should load into each well). The recognizers, labeled with fluorescence, are added, and they bind to the N-terminal amino acid. How long the recognizer stays bound is based on specificity and affinity. This generates a specific signal as the fluorescence intensity, lifetime, and binding kinetics are measured on an integrated semiconductor chip. Once the signal is captured, the terminal amino acid is cleaved, and the process repeats for the next amino acid.
Early users of Quantum-Si’s Platinum platform include scientists in a structural proteomics laboratory headed by Stephen Fried, PhD, an assistant professor of chemistry at Johns Hopkins University. The laboratory’s work encompasses whole proteomes. For example, the laboratory studies protein folding on a proteome-wide scale.
At the laboratory, graduate student Edgar Manriquez-Sandoval has started working with the Platinum. He is still in the testing stages, but he is quick to emphasize the importance of having good sequencing when studying peptides. Currently, the laboratory uses MS to sequence the peptides, but it is evaluating whether the limitations of MS can be overcome with Platinum, especially in unconventional studies. The laboratory is working with Platinum to develop new chemistries to enable full-length sequencing with single-molecule resolution.
Manriquez-Sandoval says that once the technology is sufficiently mature, it will enable very sensitive experiments and allow the laboratory to study proteins that are in lower abundance. He believes that this will open up the “dark side of the proteome.”
MS will be around forever, Manriquez-Sandoval remarks, but he speculates that the Platinum will tackle questions that have eluded MS and other technologies. The Platinum is often pitted against MS, he notes, but what matters is what the technology can give you, which is information about the sequence.
The next hurdle, Manriquez-Sandoval says, is reconstructing the sequence from the data. Here, it is unfair to compare Platinum to MS, which has had 70-plus years of development. The Platinum, and any protein sequencing platform, yields a brand-new form of data. Accordingly, Manriquez-Sandoval expects that data analysis will be challenging: “We don’t quite speak the language yet.”
From DNA to protein
One of the companies in (or moving into) protein sequencing has deep roots in DNA sequencing. When GEN asked Oxford Nanopore Technologies (ONT) for an update, we were pointed toward last year’s London Calling, ONT’s annual conference. The last section of the “Update from ONT” session, given by Clive Brown, the company’s CTO, was dedicated to protein sequencing.
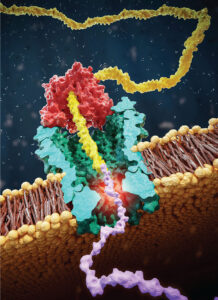
He said that ONT has a “very active program working on protein sequencing.” The goal is to use the same platform used to sequence DNA for protein. However, he admits that proteins are more difficult to get through a pore because they are folded, may not be charged, and can be heavily modified. The company’s approach involves capturing a double stranded DNA–peptide conjugate (the peptide is flanked by two DNA handles.) The DNA motor ratchets the peptide through the pore and the peptide is read. He said that getting the peptide signal has been “quite a struggle, but we’ve got it.” The core hard problem, he explains, is generating a “sequence specific signal for a peptide that is reproducible.”
Their goal is to take a complex mixture of proteins from a sample, digest them with a protease, create the DNA–peptide conjugate, and shotgun sequence the peptides. They want any lab biologist to be able to take a complex sample, digest it, process it like DNA, and run it on the same hardware that they use for DNA. No special box, no accessory equipment. And, he says that the process of building this technology has started like it did with DNA, and that they are on the “first rung on the ladder.”
A crowded field
Encodia, a protein sequencing company based in San Diego, was co-founded in 2015 by Mark Chee, PhD, and Kevin Gunderson, PhD. Chee, who also co-founded Illumina and once served as director of genetics research at Affymetrix, is now Encodia’s president and CEO. Gunderson, who spent almost two decades at Illumina and Affymetrix, is a scientific advisor at Encodia. (At the company’s founding, he was the chief technology officer.)
Encodia came out of stealth in early 2021. In a 2020 interview, Gunderson told GEN that Encodia “wants to be to the proteomics field [what] Illumina was to genomics.” He confided that Encodia was developing technology that involves “reverse translating proteins back into a DNA code that you can then read out on NGS.”
Nautilus Biotechnology is based in Seattle, WA, and it opened an office in San Diego, CA, earlier this year. At a seminar session conducted in December 2022, as part of the HUPO World Congress in Cancun, Mexico, Parag Mallick, PhD, founder and chief scientist at Nautilus, shared his frustration that genomics should continue to outpace proteomics. He also described his company’s technology, and how it employs a sample prep procedure that immobilizes individual, intact protein molecules on a large, hyperdense array to get to single molecules. The proteins are denatured and labeled with a chemical reagent to attach them to a nanoparticle scaffold. Click chemistry creates DNA-protein scaffold conjugates. The proteins are then probed, repeatedly, with multi-affinity probes while images are taken. Using multi-affinity probes targeting short epitopes, the company’s instruments iteratively interrogate the samples, and a machine learning utility identifies and quantifies the proteoforms.
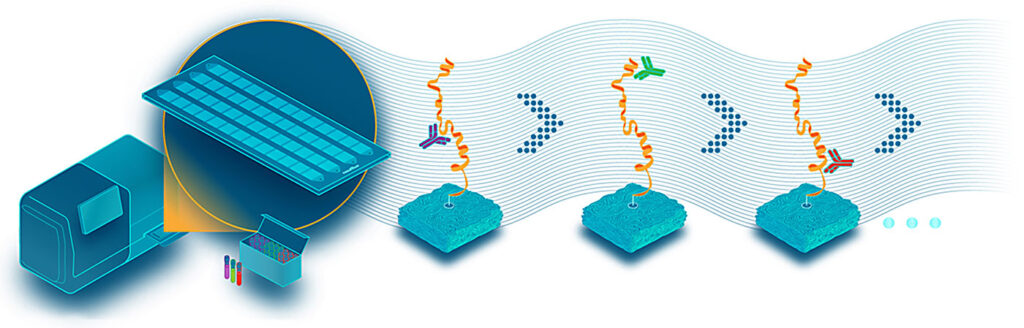
The company’s scientists compare their technology to Guess Who?—a game in which players ask a series of general questions about a person’s appearance to collect answers that can be used to determine the person’s identity. The technology involves the building of affinity reagents that can be used to target short epitopes and, ultimately, generate patterns that can allow proteins to be identified. The company notes that this method, which is called Protein identification by Short epitope Mapping (PriSM), can identify more than 95% of the proteome. Protein sequencing is just one of a number of technologies that Nautilus Biotechnology is using to explore the proteome. See our Close to the Edge interview with the company for more information.
Whether it is considered a form of social ascendancy or likened to a race, protein sequencing is just beginning. And more participants are arriving. However, not all of these companies are eager to publicize their efforts. Indeed, the representatives of one prospective protein sequencing company said that a discussion of its activities would be premature. In any event, given the complexities of the proteome, the advances in proteome-probing technologies, and the biological insights to be gained about the former through application of the latter, the vying for status—or the race—will be exciting to watch.