Scientists say they have greatly simplified the method for cell-free protein synthesis (CFPS), a technique that could become fundamental to medical research. CFPS provides the novel ability to biosynthesize proteins in a test tube in a matter of hours without the need for living cells. The approach provides a new level of control over protein production for researchers pursuing high-throughput testing, biosensor construction, metabolic engineering, and more, according to the team from Cal Poly, San Luis Obispo.
“This biotechnology harnesses the genetic code in a test tube, providing direct access to biological machinery that is traditionally locked inside the cell,” said Javin Oza, PhD, a biochemistry professor. “This allows scientists and engineers to make vaccines and therapeutic proteins, and perform diagnostic tests on-demand in the lab or out in the field. In the classroom, CFPS allows students to learn about the genetic code in an inquiry-based manner.”
While the acceptance of CFPS as a promising technology has grown substantially in the past two decades, it still suffers from some limitations due to the difficulty and expense of implementing the technique.
As reported (“Escherichia Coli-Based Cell-Free Protein Synthesis: Protocols for a robust, flexible, and accessible platform technology”) in the Journal of Visualized Experimentation, a group led by Oza and collaborator Katherine Watts, PhD, has developed a method to make CFPS widely accessible. The main advantages of the new technique are speed, cost-effectiveness, and a much less complex reaction setup compared to other CFPS systems, noted Oza, who added the publication includes a video guide for implementing the new procedure.
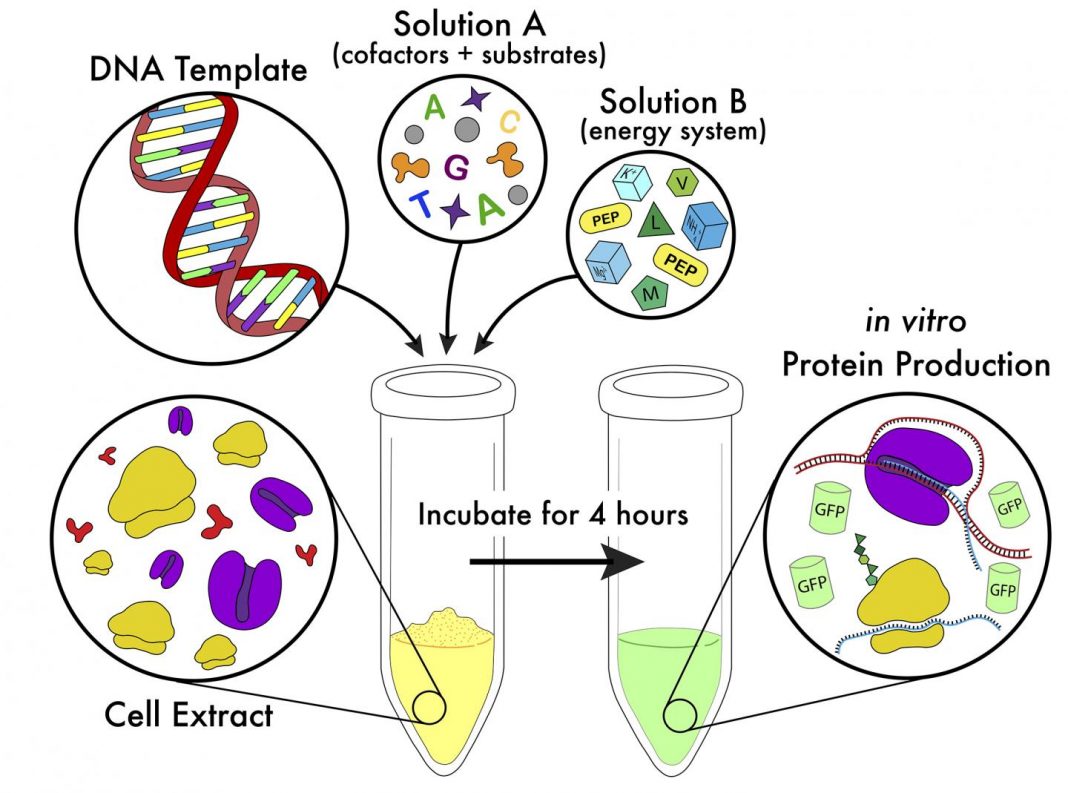
“Over the last 50 years, CFPS has emerged as a powerful technology to harness the transcriptional and translational capacity of cells within a test tube. By obviating the need to maintain the viability of the cell, and by eliminating the cellular barrier, CFPS has been foundational to emerging applications in biomanufacturing of traditionally challenging proteins, as well as applications in rapid prototyping for metabolic engineering, and functional genomics. Our methods for implementing an E. coli-based CFPS platform allow new users to access many of these applications,” the investigators wrote.
“Here, we describe methods to prepare extract through the use of enriched media, baffled flasks, and a reproducible method of tunable sonication-based cell lysis. This extract can then be used for protein expression capable of producing 900 µg/mL or more of super folder green fluorescent protein (sfGFP) in just 5 h from experimental setup to data analysis, given that appropriate reagent stocks have been prepared beforehand. The estimated startup cost of obtaining reagents is $4,500 which will sustain thousands of reactions at an estimated cost of $0.021 per µg of protein produced or $0.019 per µL of reaction,” the investigators wrote.
“Additionally, the protein expression methods mirror the ease of the reaction setup seen in commercially available systems due to optimization of reagent pre-mixes, at a fraction of the cost. In order to enable the user to leverage the flexible nature of the CFPS platform for broad applications, we have identified a variety of aspects of the platform that can be tuned and optimized depending on the resources available and the protein expression outcomes desired.”
“This protocol simplifies and clarifies the methods for implementing cell-free protein synthesis by non-experts,” Oza said. “Improved access to these methods will help democratize the platform and its broad set of applications.”
The new technique requires only basic laboratory training for new users to implement CFPS in their labs, from cell-growth and extract preparation to the in vitro protein synthesis reactions themselves, he continued.
“In our collaboration, we are employing the simplified CFPS method to synthesize and engineer complex mega-enzymes, which are traditionally difficult to express,” said Watts, also a Cal Poly biochemistry professor. “The use of CFPS allows for much more rapid design-build-test cycles for engineering these mega-enzymes.”

