We’re used to the idea that we might be identified through our unique DNA signatures. But now we may have to become accustomed to yet another way of being singled out on the basis of DNA. The DNA, it turns out, doesn’t have to be our own. Instead, it can belong to our microbiomes—the microbial communities we carry in and on our bodies. Microbiomes, a new study demonstrates, have a personal dimension. They contain so many distinguishing features that it is possible for individuals to be uniquely identified among populations of hundreds.
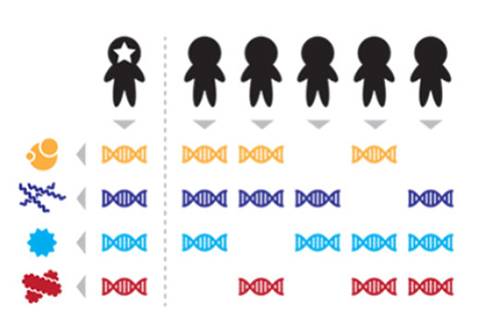
The new study was carried out by scientists at the Harvard T.H. Chan School of Public Health. Aware that person-to-person microbiome variations vary, and that many of these variations persist over time, the Harvard scientists decided to find out if there were such a thing as a microbiome “fingerprint.”
The Harvard scientists used publicly available microbiome data produced through the Human Microbiome Project (HMP), which surveyed microbes in the stool, saliva, skin, and other body sites from up to 242 individuals over a months-long period. Then the scientists evaluated the data using a “set-based coding algorithm.” Essentially, the scientists combined stable and distinguishing sequence features from individuals’ initial microbiome samples into individual-specific codes. They then compared the codes to microbiome samples collected from the same individuals’ at follow-up visits and to samples from independent groups of individuals.
The scientists published their work May 11 in the Proceedings of the National Academy of Sciences, in an article entitled, “Identifying personal microbiomes using metagenomic codes.”
“Our results demonstrated that individuals could be uniquely identified among populations of 100s based on their microbiomes alone,” the authors wrote. “In the case of the gut microbiome, >80% of individuals could still be uniquely identified up to a year later—a result that raises potential privacy concerns for subjects enrolled in human microbiome research projects.”
“Linking a human DNA sample to a database of human DNA ‘fingerprints’ is the basis for forensic genetics, which is now a decades-old field. We’ve shown that the same sort of linking is possible using DNA sequences from microbes inhabiting the human body—no human DNA required,” said Eric Franzosa, Ph.D., a research fellow in the Department of Biostatistics at Harvard Chan. “This opens the door to connecting human microbiome samples between databases, which has the potential to expose sensitive subject information—for example, a sexually-transmitted infection, detectable from the microbiome sample itself.”
“Although the potential for any data privacy concerns from purely microbial DNA is very low, it’s important for researchers to know that such issues are theoretically possible,” said the PNAS article’s senior author Curtis Huttenhower, Ph.D., associate professor of computational biology and bioinformatics at Harvard Chan. “Perhaps even more exciting are the implications of the study for microbial ecology, since it suggests our unique microbial residents are tuned to the environment of our body—our genetics, diet, and developmental history—in such a way that they stick with us and help to fend off less-friendly microbial invaders over time.”