
NanoString
Spatial multiomics, still in its discovery phase, can mean different things to different people. It typically denotes the visualization of transcriptomic and proteomic data in the context of tissue architecture, either directly on the same section or on serial sections that are integrated computationally.
Spatial multiomics may eventually grow to encompass lipids, glycans, metabolites, epigenetic markers, and transient post-translational stamps on proteins. “Every new technology in medicine grows from discovery, through translation, to diagnostics,” says Joachim Schmid, PhD, vice president, R&D spatial informatics & AI, NanoString Technologies.

U. Ill. Urb.-Champ.
Yet, even in its current incarnation, spatial multiomics is being used in pathology research laboratories to establish precise methods of identifying and classifying diseases, and in determining the specificity and efficacy of drugs. According to Jonathan Sweedler, PhD, the James R. Eiszner Family Endowed Chair in Chemistry at the University of Illinois at Urbana-Champaign, “For some classes of molecules, spatial multiomics can already get quality chemical distributions within tissues and tumors. Mass spectrometry imaging and vibrational spectroscopy provide molecular information related to tissue health.”
Immunotherapy has ushered in the need for meticulous and reproducible molecular analyses for the granular classification of diseases that make it possible to match patients to optimal therapies.

Nucleai
“Things are getting complicated for the pathologist. It used to be simple—look at a few H&E and IHC slides, pass it on to the molecular lab—but now we want to know the relationship of tumor microenvironments (TMEs) to tumor cells. That’s where spatial comes in,” says Kenneth Bloom, MD, head of pathology, Nucleai. “We must identify cell types in the TME at the same time and understand their relationships because sometimes cells only work in certain ways when they are adjacent to other cells.”
Elizabeth Neumann, PhD, an assistant professor of chemistry at the University of California, Davis, states, “Diseases that we once thought to be homogenous can be classified into subtypes as we get more spatial multiomics data.”
At Pathology Visions 2022 (a conference that was held last October in Las Vegas, NV), Schmid sensed excitement around visualizing single molecules in spatial contexts. Digital pathology digitalizes entire specimens on glass slides through whole-slide imaging and uses virtual microscopy and computational methods to uncover clinical insights. “This was the first time that the conference was sold out,” Schmid notes. “Thought leaders are talking about getting into the spatial biology space. Nobody yet knows how it will be translated into daily work, but spatial multiomics generates a lot of digital information which fits the field.”
Pathologists have historically relied on formalin-fixed paraffin-embedded (FFPE) tissue. Spatial technology is being developed for FFPE, as well as for fresh-frozen tissue. Nevertheless, overhauling legacy technologies for complex workflows requires solid benefits.
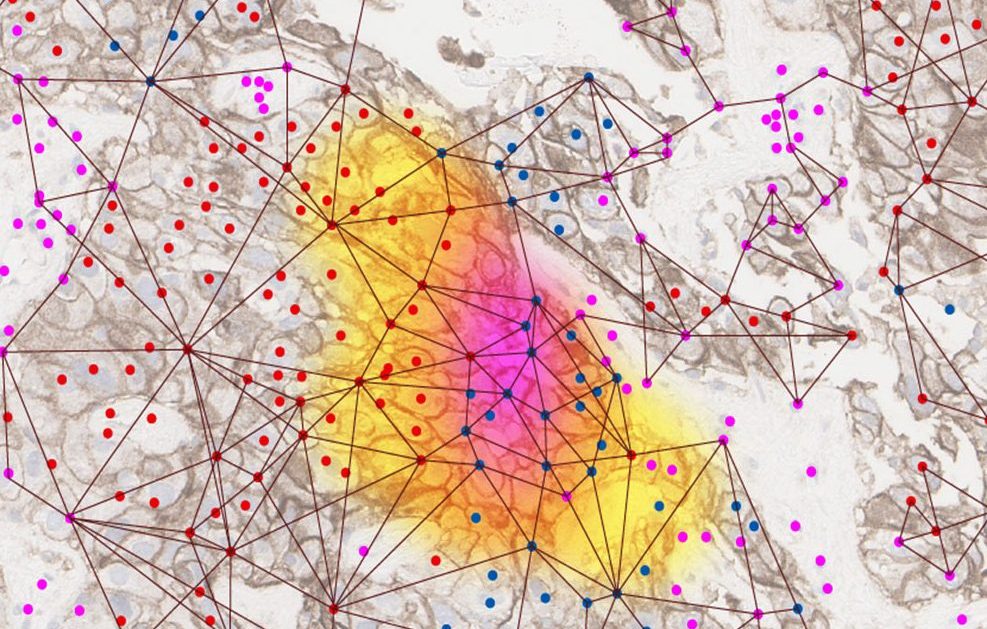
“If you look at a tissue under a microscope with your eyes, the amount of information you get is limited,” Sweedler remarks. “Pathologists have been innovative at staining tissue so that the molecules of interest are visible to infer disease states. These work. The question is whether it is possible to get more granular information through spatial multiomics.”
Benefit to patients is the ultimate driver for the implementation of new technologies. The use of spatial multiomics offers distinct advantages for patient stratification, not only in acquiring more information to validate preliminary results, but also in acquiring information that could not be accessed earlier.

Akoya Biosciences
“You can use a combination of proteins as a [disease] signature, but in many cases to understand the mechanism of the treatment or disease progression, it may be advantageous to have a cell surface protein together with cytokines and chemokines that can be much better measured through RNA,” says Julia Kennedy-Darling, PhD, vice president of innovation, Akoya Biosciences.
In some instances, spatial multiomics is an alternative validation, but in others it is the only route to answers. “If you have high Her2 RNA and protein, it gives you confidence. The advantage is apparent when both have medium to low expression,” Kennedy-Darling says. Analyzing different classes of biomarkers sequentially—as is the current practice—to arrive at diagnostic decisions takes time, whereas acquiring integrated datasets on the same tissue section allays doubt and increases speed. On the other hand, to identify the source of secreted proteins such as cytokines, it is imperative to detect RNA since the protein is dispersed.
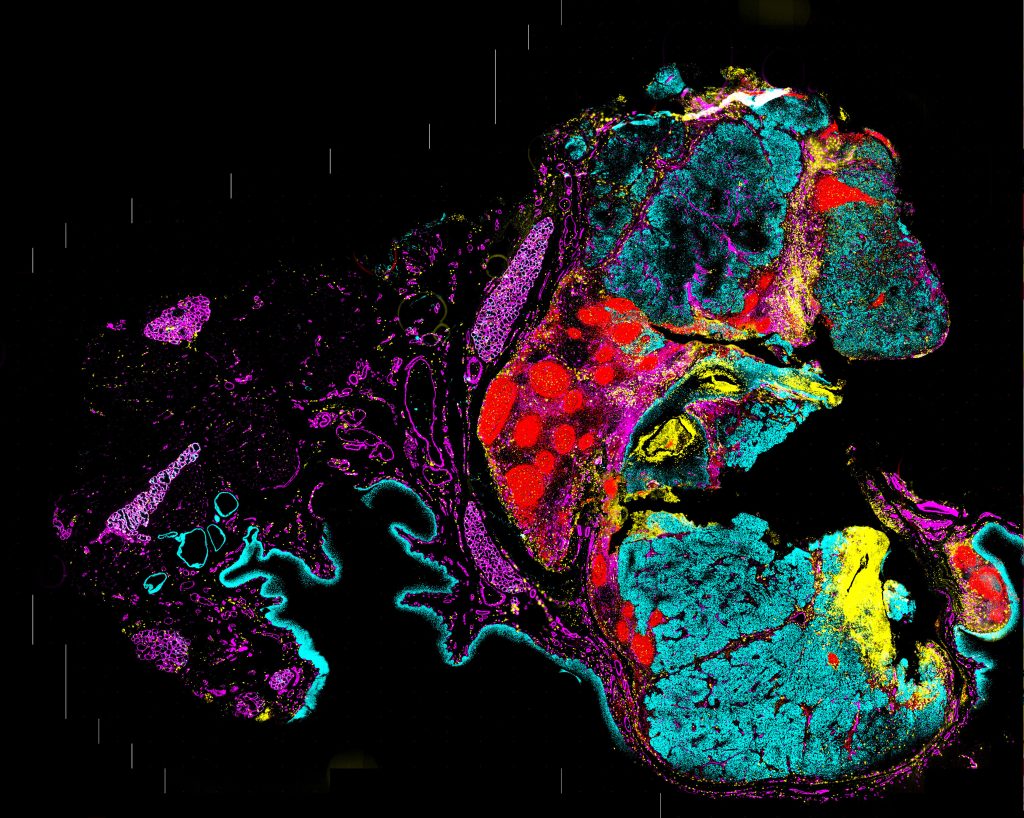
Location, heterogeneity, and temporal landscapes
By determining the sources and destinations of secreted proteins and the receptors they bind, it is possible to map the temporal landscape of signaling pathways within the spatial context.
“Most spatial technologies have focused on delivering a detailed snapshot of what exists. To be able to layer on how the signaling networks are perpetuating spatially is profound,” Kennedy-Darling continues. Such spatiotemporal approaches can provide concrete evidence of cellular interactions and signaling networks. Until now, investigators have had to rely on tenuous correlations.

Johns Hopkins Univ.
Significance in biology, like value in real estate, is largely determined by location. Yet, the power of understanding spatial relationships between cells has been lost in most molecular studies.
Predictions of responses in immune oncology are far better when assays include a spatial or co-expression component than when they rely on the presence or absence of a marker, notes Janis Taube, MD, professor of dermatopathology, Johns Hopkins University. In addition to tipping the scales toward higher predictive value, spatial multiomics offers quantifiable measures of cellular heterogeneity. “Pathologists know tumors are heterogenous,” Taube notes, “but being able to put numbers to that is going to be an exciting new development in prognosis and prediction, hopefully for both discovery research and clinical pathology.”
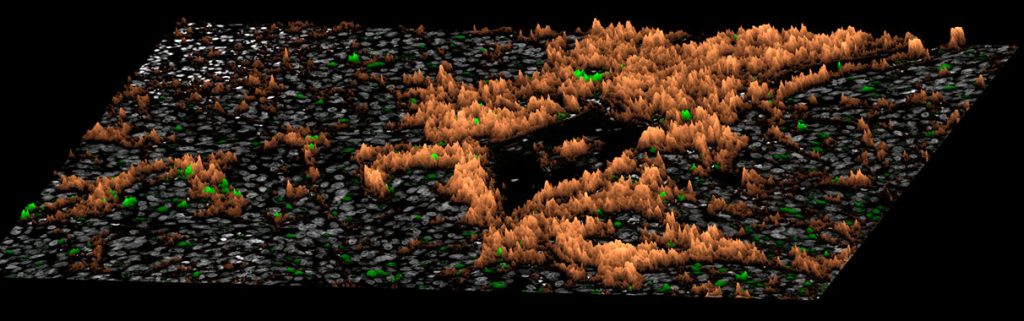
Precision oncology
Tumors consist of a dizzying array of cancer and immune cells with unique molecular signatures. Single-cell spatial multiomics that computationally integrates morphological data with genomic, transcriptomic, and proteomic biomarkers have been transformative in uncovering markers for the heterogenous TME, tumor growth, metastasis, and drug resistance. Precision oncology rests on the ability of pathologists to identify cancer cell subtypes and TME characteristics to improve diagnosis and treatment.
“As we were staining tumors with multiple immunofluorescent markers, we were running into issues of data analysis, curation, and storage as well as issues of normalization and calibration,” Taube recalls. “Astronomy dealt with similar issues 25 years ago, when scaling up and scaling out multispectral images. We took our lessons from the Sloan Digital Sky Survey and applied them to pathology specimens of tumors.”
Taube’s team has developed a platform called AstroPath to develop biomarkers for patients with advanced stages of cancer and is currently focusing on earlier stages.
In a recent paper, investigators used Insilico Medicine’s AI-powered multiomics platform, PandaOmics, to explore gene expression changes in rare DNA repair-deficiency disorders. The analysis uncovered a scaffolding protein (CEP135) important in cell division that was depleted in DNA repair diseases with a high predisposition for cancers. Stratifying patients based on CEP135 expression enabled the investigators to identify a potential therapeutic target (PLK1) for sarcoma patients. Although clinical validations remain pending, the study highlights the power of spatial multiomics in the rapid discovery of biomarkers and therapeutic targets.

UC Davis
Spatial multiomics may also help ascertain tumor margins before and after treatment. “How you define the boundaries between healthy tissue and cancer is not trivial,” Neumann stesses. “With traditional immunofluorescence, on a good day, you can detect a maximum of seven markers. You cannot even label all immune cells with that.” Although FFPE retains most spatial information, some molecules are too few or labile to be detected, attracting novel approaches of tissue processing and image analysis.
“Our technology is developed for FFPE—the standard in pathology practice. It also works on fresh-frozen samples. The research question ultimately determines the sample type and analytical tools you use,” Schmid advises. “Sensitivity and data quality is one piece of the puzzle in detecting low-abundance proteins and RNA. The other piece is data analysis and quality control of the data.”
“FFPE biobanks are an incredible resource, but fresh-frozen tissue is sometimes easier to use for molecular information–rich mass spectrometry,” Sweedler details. “For each category of biomolecules, processing steps have to be worked out separately and optimized.” Sweedler, whose work involves unstable neuropeptides, has found that laser heating deactivates degrading enzymes rapidly and is an efficient method for tissue stabilization. “Limitations should not discourage you,” Sweedler insists. “These are opportunities for advancement.”
Whereas spatial multiomics on the same and serial sections of FFPE or fresh-frozen tissue are both helpful, each has its own merits and challenges. “The advantage of serial section multiomics is in the established workflows. The challenges are in data analysis,” Kennedy-Darling clarifies. However, when different molecules are imaged on different sections and combined, a difference of one cell between sections can lead to errors. “With the same section, you are measuring the targets directly,” she adds. “There is no question about the localization of targets.”
Data dilemma
Spatial multiomics yields big data, creating challenges in data storage, sharing, accessibility, and analysis, in addition to the challenge of re-equipping the pathology workforce with software skills.
“You want to generate the data and store it in a well-organized fashion at a single location. You can then generate web browsers that overlay the data and allow queries on the database from anywhere in the world,” Taube notes.
“Our AtoMx spatial informatics platform will support the analysis and review of the information,” Schmid asserts. “The cloud enables you to work without worrying about storage and allows easy access [to data] among collaborators.” Schmid believes it’s important to create customized data analysis solutions based on the users’ needs. While some need basic methods and established pipelines, others want to import open source algorithms or code their own methods. “There isn’t one way that’ll be able to provide all you need. We want to create an ecosystem that allows users to connect the dots,” Schmid relates.
Different laboratories store similar data in unique formats, which poses an obstacle for comparative studies. Few standards exist to facilitate cloud computing. Companies and academics support standardization efforts by proposing standards for each step in the pipeline, adhering to popular formats or working through committees such as the one responsible for the Digital Imaging and Communications in Medicine standard.
“Standardization is powerful, but it is a slow process. The challenge is to not limit innovation,” Schmid declares. NanoString supports the OME-TIFF format for imaging data, and Seurat for “weighted-nearest neighbor” analyses.
The ability of spatial multiomics to combine diverse molecular and morphological information in analyzable formats aligns it with current trends in pathology. Although its discovery phase involves analyzing large numbers of biomolecules in research pathology laboratories, translational studies will likely funnel unwieldy panels into a few salient markers that encapsulate the clinical crux to benefit diagnosis and precision medicine. Ultimately, technology does not determine its application—necessity remains the uncontested mother of innovation.

Comments are closed.