The spatial distribution of cell surface proteins, which governs vital processes of the immune system such as inter-cell communication and mobility, has proven difficult to assess. New tools are needed that not only capture spatial organization of immune cells, but also multiplex at a high level while delivering high resolution and throughput.
Flow cytometry using fluorophore-labeled antibodies has been extensively used to study proteins on immune cells for several decades. More recently, efforts have been made to overcome the multiplexing limitations of conventional flow cytometry by instead labeling antibodies with isotopes for mass spectrometry readout, or with oligonucleotides for next-generation sequencing readout.
Although these approaches can be used to characterize and phenotype cells at high multiplex and throughput, the information they provide pertains only to the abundance of each target protein on each cell. They do not describe the spatial organization of the targeted molecules.
Fluorescence microscopy has traditionally been used to study the spatial organization of proteins on single cells, but multiplexing is limited to a few targets due to the spectral properties of fluorophores, and the signal-to-noise ratio suffers from autofluorescence and spectral bleed-through between channels. Furthermore, the view provided by each microscopy image is limited to a selected focal plane, so if the whole cell surface is to be represented, a Z-stack of images for each fluorophore is required, limiting throughput.
Recently, methods solely relying on oligonucleotide sequences to image biological samples have been demonstrated. Sometimes referred to as “DNA microscopy,” these methods rely on the incorporation of DNA tags that can be decoded to reveal both biomolecule identity and position within the biological sample. These methods offer possibilities to circumvent the limitations in multiplexing, throughput, and (potentially) resolution that beset optical imaging–based methods.
Pixelgen Technologies has developed Molecular Pixelation (MPX) technology to unlock a new spatial dimension to single-cell proteomics research by supplementing abundance information with spatial information about target proteins. This added spatial dimension provides researchers with opportunities to gain deeper insights into cell function at sub-cellular resolution.
The MPX protocol can be performed using standard molecular biology laboratory equipment, without the need for any dedicated hardware or consumables to compartmentalize cells, and a dedicated data processing pipeline is available for DNA processing and analysis of the sequencing output. The reagent kit contains an 80-plex panel against cell surface receptor targets on the major types of peripheral blood mononuclear cells (PBMCs)—T cells, B cells, natural killer cells, and monocytes—and allows for sequencing of up to 1,000 cells per sample and a total of eight samples per reagent kit. Dedicated data processing software tools are available for straightforward data processing and analysis of the rich data that the technology generates.
MPX workflow overview
The MPX workflow can be divided into six steps: a cell preparation step, two pixelation steps, an NGS preparation step, an NGS step, and an analysis step (Figure 1). During the cell preparation step, the immune cells in suspension are chemically fixed with paraformaldehyde to lock the surface proteins in place and prevent any reorganization during downstream sample processing. The fixed cells are blocked, and a target panel of 80 antibody-oligonucleotide conjugates (AOCs) is added, whereupon the AOCs bind their surface receptor targets. Next, the pixelation steps consist of serially hybridizing a set of so-called DNA pixels to the oligonucleotide portion of AOCs bound to cells. DNA pixels are single-stranded DNA molecules produced by rolling circle amplification, where each unique DNA pixel molecule contains repeats of a unique sequence identifier. Each DNA pixel molecule can hybridize to multiple AOCs in proximity on the cell surface.
![]()
The DNA pixel identifier sequence is then incorporated onto the hybridized AOC via a gap-fill ligation enzymatic reaction, forming about 1,000 neighborhoods on the cell surface where all AOC molecules within each neighborhood now share the same DNA pixel identifier sequence. The hybridization and gap-fill ligation reactions are then repeated for a total of two pixelation steps, thereby creating two sets of partially overlapping neighborhoods across the cell surface of each assayed cell.
Each generated amplicon contains a protein identifier barcode, a unique molecular identifier sequence, two DNA pixel identifier sequences, and PCR primer sites. The generated amplicons are finally amplified by PCR, purified, and quantified for Illumina sequencing.
Data processing and spatial inference
In short, the dedicated data processing pipeline, which is called Pixelator, receives the sequencing reads and subjects them to quality filtering, decoding (to establish protein identities), error correction, and consolidation (to collapse identical reads into unique sequences). Each sequenced unique molecule can be represented as an edge (link) of a graph (network) with the DNA pixel identifier sequences as nodes and the protein identity tags as edge or node attributes. Separated “cell graphs” representing individual cells are contained within the sample-level graph generated from a sequenced sample.
Spatial inference of the relative locations of individual AOC molecules is possible by interrogating the relative positions of the AOCs within each cell graph. This also allows for the calculation of spatial metrics such as the degree of clustering (polarity) of each of the 80 protein targets, or the level of colocalization between pairs of protein targets.
Results
Data analysis of protein abundance can be performed on MPX data similarly to other multiplexed single-cell methods. For example, PBMCs taken from a healthy donor were processed through the MPX protocol, and then a uniform manifold approximation and projection (UMAP) dimensionality reduction was performed on the protein count matrix output, which formed separated clusters that were consistent with the expected protein signatures for the major cell types expected in the PBMC samples (Figure 2). The fraction of each cell type was also consistent with expected fractions seen in healthy PBMC donors.

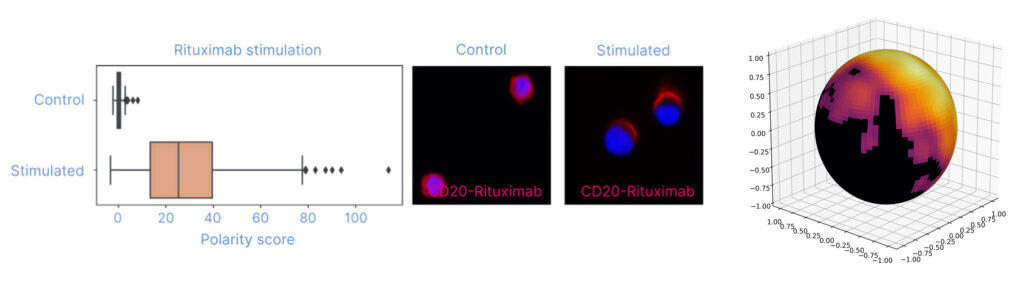
To demonstrate the added spatial dimension of the data, Raji B cells were treated with an AOC of the CD20 therapeutic antibody drug rituximab before fixation and processing of the treated cells and untreated control cells through the protocol. Rituximab is known to cluster CD20 on B cells, which should then be reflected in the rituximab polarity score output of data.
The clustering of CD20 occurring upon rituximab AOC treatment was confirmed with fluorescence microscopy (Figure 3). Polarity scores for rituximab depicting the degree of clustered protein expression were compared between stimulated and control samples, and they showed a significant elevation of polarity scores for rituximab-treated cells. Additionally, graph representations of individual rituximab-treated cells, colored by the count density of rituximab of each node, showed a clustered expression pattern consistent with microscopy validation.
Conclusion
Unlocking a new spatial dimension to single-cell proteomics research at high multiplex and throughput can enable researchers to gain additional and deeper insights into immune cell function at scale. Example data from Pixelgen Technologies’ MPX technology showcases the ability to detect differential spatial clustering of a target protein confirmed to be clustered upon stimulation with rituximab.
Filip Karlsson is co-founder and chief technology officer of Pixelgen Technologies. Website: www.pixelgen.com.


