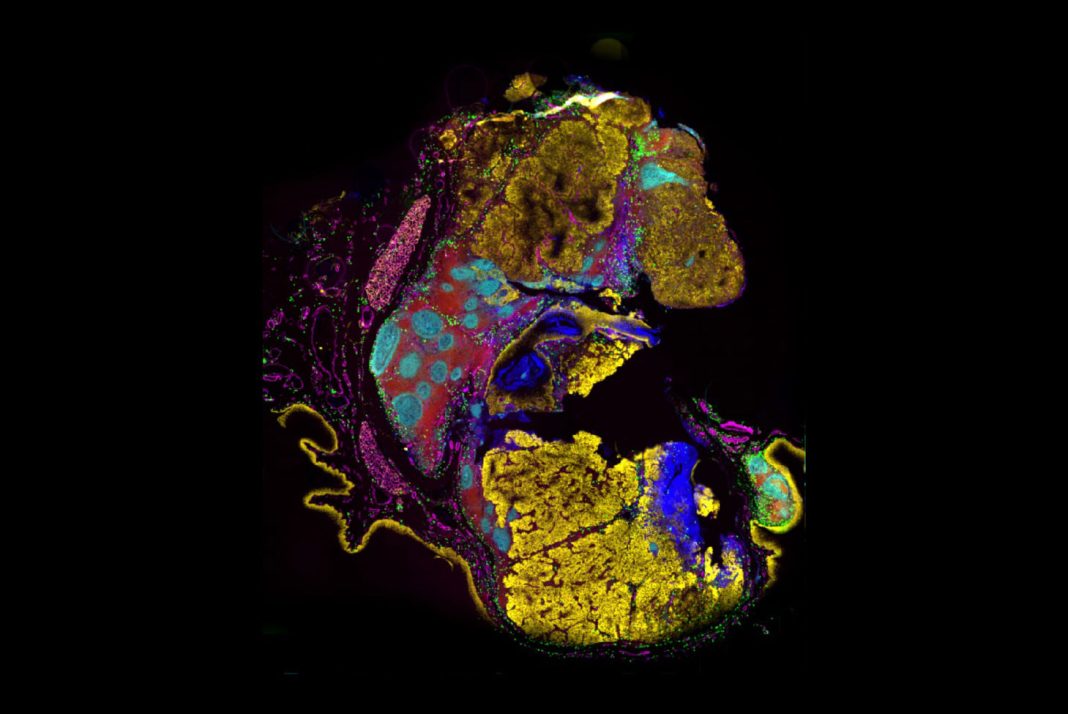
Current high-plex approaches for studying single-cell biology, such as flow cytometry and single-cell RNA sequencing, reveal the cell populations present in a sample but are unable to characterize the spatial organization and interactions between cells. Meanwhile, traditional low-plex immunohistochemistry (IHC) informs scientists about how cells localize but lacks the level of information needed to capture the complexity of the tissue microenvironment.
To perform in-depth, unbiased analysis, scientists require ultrahigh-plex detection methods that preserve spatial context. “NGS-based single-cell data provide crucial information about the cell types and functional states present in your samples, but it doesn’t tell you where they are located in tissue,” says Oliver Braubach, PhD, Head of Research Applications at Akoya Biosciences. “Spatial context is essential for determining how cells organize and interact. Understanding the spatial interactions between cells will help us uncover novel biomarkers and disease mechanisms.”
Scientists are realizing the importance of studying cells in context, and the tools that uncover the spatial distribution and relationships between cells will fuel the next wave of biological discovery.
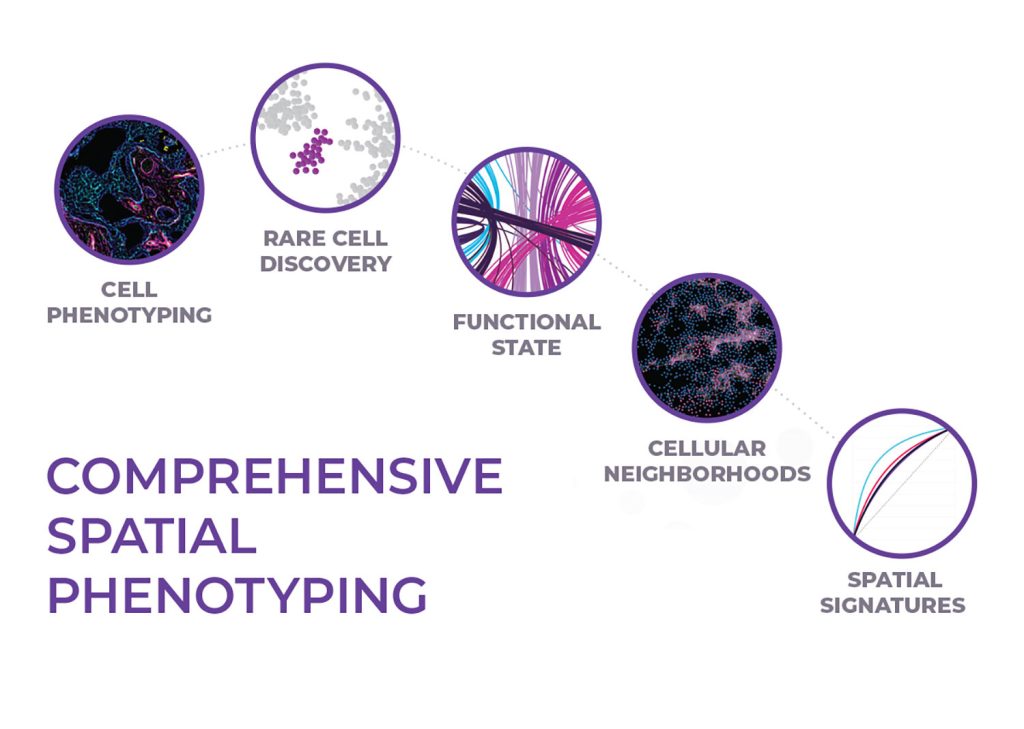
Spatial phenotyping enables unbiased discovery
The true power of spatial biology lies in phenotyping every cell across whole slides, an approach known as spatial phenotyping. Spatial phenotyping enables scientists to visualize and quantify hundreds of biomarkers in a single tissue sample while maintaining cellular and sub-cellular detail. This approach enables rare cell detection, identification of cellular neighborhoods, and ultimately aids in developing predictive spatial signatures.
“It’s only when one actually sees and accounts for every cell in a sample that we can begin to appreciate all of the details that could be missed,” says Braubach.
This approach is what powers Akoya’s multiplex imaging platforms. The PhenoCycler™ solution (formerly CODEX®) uses an iterative imaging workflow to reveal 100+ biomarkers across whole tissue sections. PhenoCycler is ideal for hypothesis-free discovery, generating large datasets that can be analyzed to identify “cellular neighborhoods.”

Cellular neighborhoods—a seminal concept in spatial biology—came from the lab of Garry Nolan, PhD, Rachford, and Carlota A. Harris Professor in the Department of Pathology at Stanford University. In a seminal paper published in Cell, Schürch et al. used PhenoCycler to develop high-dimensional spatial maps of colorectal cancer (CRC) tissue. They not only identified distinct cell types but revealed how these cell types are organized into distinct niches and neighborhoods. They showed how a cellular region’s ability to organize into structured neighborhoods was associated with less lethal disease, while diffuse cellular structures were associated with more lethal forms of disease.
Cellular neighborhood analysis is also being compiled in the Human Cell Atlas. Kai Kessenbrock, PhD, Co-Principal Investigator of the Human Breast Cell Atlas Project, is taking advantage of PhenoCycler’s ultrahigh-plex capabilities in combination with single-cell transcriptomics to build a spatially resolved, single-cell, multiomic map of human breast tissue.
Spatial biology 2.0: Scaling up discoveries by virtue of speed
Akoya Biosciences recently launched the PhenoCycler-Fusion system that combines the strengths of the company’s two foundation technologies. Capable of mapping 1 million cells in just 10 minutes, the system integrates the ultrahigh-plex PhenoCycler workflow with the powerful, high-throughput imaging capabilities of the company’s PhenoImager™ (formerly Phenoptics™) solution.
This breakthrough solution delivers true whole-slide, unbiased imaging in minutes at greater level of multiplexing empowering users to scale up spatial discoveries. Equipped with PhenoCycler-Fusion, Akoya’s applications team led by Dr. Oliver Braubach has demonstrated how this solution can redefine spatial imaging. With PhenoCycler-Fusion, deep ultrahigh-plex spatial discovery has now become a standard at Akoya. Meticulously designed 100-plex antibody panels have proven invaluable for uncovering the mechanistic insights into tumor biology. These kinds of deep investigation aid in revealing mechanisms underlying clinical response and therapeutic resistance. Conventional methods for single-cell biology studies are simply not equipped for simultaneous detection of intersecting predictors. With PhenoCycler-Fusion, however, the ability to simultaneously study multiple facets of tissue biology—ranging from metabolomics, proliferation, and stress response to the immune microenvironment—is now within reach.
Along with its well-established protein biomarker assays, the PhenoCycler-Fusion will also enable RNA detection from the same sample to enable a more comprehensive exploration of tissue biology.
Through high-speed, ultrahigh-plex, and multiomic analysis of whole slides at single- and sub-cellular resolution, PhenoCycler-Fusion is setting a new standard in spatial biology.
From spatial discoveries to spatial phenotypic signatures —introducing the next class of biomarkers
Akoya’s high-throughput (HT) instrument, PhenoImager HT, enables fast, whole-slide imaging at single-cell resolution. The PhenoImager HT workflow utilizes focused panels for high-throughput biomarker validation of spatial biomarker signatures and can process 300+ samples per week.
In a study published in the Journal of the American Medical Association (JAMA), Lu et al. found that spatial phenotypic signatures, identified using multiplex immunofluorescence (mIF), had much higher predictive power compared to other approaches, such as single-plex PD-L1 IHC or gene expression profiling.
Akoya has partnered with AstraZeneca to advance new spatial biology workflows and spatial phenotypic signatures based on the PhenoImager workflow. Together, the two companies aim to develop and implement predictive assays and analysis frameworks to enable drug development informed by spatial biomarkers. PhenoImager is also the foundation for Johns Hopkins’ AstroPath™ platform for deep spatial profiling of tumor sections, developed jointly by the university’s astrophysics and pathology departments. In a Science paper, Berry et al. describe how they used AstroPath to build two-dimensional maps of the tumor microenvironment in metastatic melanoma and identify a highly predictive spatial phenotypic signature for anti-PD-1 immunotherapy response and patient outcomes.
“High-plexing, spatial context, and improved accuracy, together extend our ability to study complex tumor micro-environments beyond standard histopathology biomarker discovery tools,” says Sneha Berry, PhD, first author of the study and Akoya Lead Scientist.
Serving the diverse needs of researchers across discovery, translational and clinical research is key. Akoya offers a full continuum of spatial phenotyping solutions emphasizing more biomarkers, more throughput, and more translation into the clinic.
To learn more about PhenoCycler-Fusion, visit us at akoyabio.com/fusion