The search for a universal flu vaccine has been long and fraught with false starts and dead ends. For more than five years, Nicholas Heaton, PhD, associate professor of molecular genetics and microbiology at Duke University, has been working with his team on a systematic approach to create such a vaccine. One particularly promising approach is to develop antibodies that target both the head and stalk of the most abundant surface protein on the influenza virus.
The group’s new study, entitled, “Vaccination with Antigenically Complex Hemagglutinin Mixtures Confers Broad Protection From Influenza Disease,” was published in Science Translational Medicine.
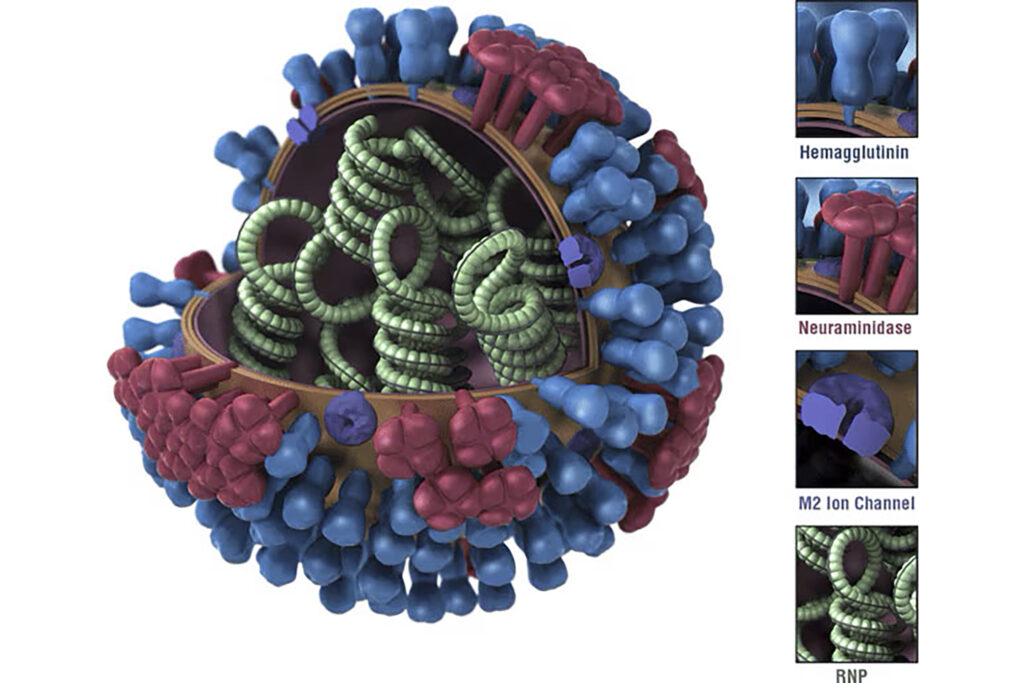
Flu is widespread and deadly, with anywhere from 9–41 million cases annually, resulting in approximately 5–51,000 thousand deaths a year in the United States, according to the Centers for Disease Control and Prevention (CDC). The severity of flu infections is as variable as the strains that cause the disease. Each strain is typically characterized and referred to by alphanumeric codes including the letters H (hemagglutinin) and N (neuraminidase), identifying the most prominent surface proteins, and numbers to delimit the protein variant. The influenza virus contains about 5–10 times more hemagglutinin than neuraminidase, said Heaton.
Current annual vaccines typically target the globular head of the lollipop-shaped hemagglutinin glycoprotein or neuraminidase proteins coating the virus. These proteins are highly mutable, resulting in the need to create new vaccines annually.
“The virus has evolved to have the immune system recognize these (features on the head region). But these are the shapes the virus can change. That is an insidious strategy,” Heaton said. Furthermore, due to the sheer quantity of flu strains, only some of them are included in each year’s flu shot.
For decades, researchers have sought out strategies to circumvent the rapid incessant evolution of flu strains. These strategies have varied from focusing on neuraminidase, to using stalk-targeting nanoparticles, or using live, genetically altered viruses.
To broaden the function of flu vaccines, Heaton’s team now reports the development of “an antigenically complex mixture of recombinant hemagglutinins designed to redirect immune responses to more conserved domains of the protein.” Asked how this approach is different from previous efforts, Heaton told GEN, “I think that all (including ours) have their pros and cons.”
As there are more hemagglutinin than neuraminidase molecules on the flu virus surface, the team focused their efforts on identifying regions of that protein that were less likely to mutate over time. “A number of groups have gone through and experimentally mutagenized the whole hemagglutinin and asked, ‘Which areas can change and still allow the hemagglutinin to function?’” Heaton explained. “The answer is, you can’t really change the stalk and expect it to continue to function.”
“Antibodies against the stalk work differently,” Heaton explained. “Their mechanism of protection is not necessarily to block the first step of infection. So our idea was: What if we can come up with a vaccine that gives us both? What if we can get good head antibodies and at the same time also get stalk antibodies in case the vaccine selection was wrong, or if there’s a pandemic?”
First, Heaton’s team aimed to create a group of mutated hemagglutinin proteins with a wide variety of mutations only in the head region, but with conserved stalks. They used gene editing techniques to create more than 80,000 variants of hemagglutinin with changes just in the head region after purification and sequencing procedures. They next verified that the mutated region was not prone to antibody recognition compared to the wildtype, suggesting that they had successfully generated a mutant protein mixture that could be used for vaccine creation.
They then tested their mixture in animal models—mice and ferrets—to determine the antibody response. “If we took your blood to see if you are likely to be protected from a strain of flu, we’d be measuring what your antibodies do to hemagglutinin as the best metric of what’s likely to happen to you. The strongest correlates of protection have to do with hemagglutinin-directed immunity,” Heaton explained.
Following this premise, the team found that animals vaccinated with the experimental mixture produced more antibodies against conserved stalk regions compared with controls. “One thing that we observed was enhanced vaccine-induced responses to both the head and stalk domain of the hemagglutinin,” Heaton told GEN. “That has been a difficult goal to achieve.”
There are many avenues of further study ahead. “One of the most important next steps will be to understand if we can apply this approach to all of the different subtypes of influenza that infect humans. In this paper, we only study H1N1 viruses,” Heaton told GEN.
Translating this data to humans will be a much larger hurdle. Heaton concluded, “There are many challenges in the advanced development of a vaccine. One major challenge is predicting how different people (with different immune exposure histories) will respond to the vaccine. That is very hard to model in a laboratory environment.”

