Prime editing uses CRISPR-guided reverse transcription to enable the programmable introduction of any desired base substitution or small insertion or deletion. One challenge for using prime editing systems is the large size of the required prime editing protein 2 (PE2). Consisting of nSpCas9 (Streptococcus pyogenes Cas9 nickase) with a mutated MMLV-RT (Murine Leukemia Virus Reverse Transcriptase) domain fused to its C-terminus, PE2 is typically 2,117 amino acids encoded by 6,351 base pairs.
Massachusetts General Hospital (MGH) researchers created a system called Split Prime Editing (Split-PE), which separated nSpCas9 and MMLV-RT proteins to function as efficiently as intact PE2 in human cells. These results and reagents have important implications for improving prime editing and should help to accelerate the optimization and application of prime editing for research and therapeutic uses.
The research article titled “Engineered CRISPR prime editors with compact, untethered reverse transcriptases” was published on September 26, 2022, in Nature Biotechnology.
The mechanics of prime editing
In existing prime editing systems, mutations are induced by a prime editing protein and a prime editing guide RNA (pegRNA). This pegRNA directs nSpCas9 activity to create an R-loop with a nicked DNA strand, which anneals to a primer binding sequence (PBS) at the 3′ end of the pegRNA. The reverse transcriptase part of the prime editing protein then reverse transcribes the reverse transcription template that is adjacent to the PBS into DNA, encoding the desired edit of interest. This DNA template then mediates the introduction of the edit into the genomic locus by a mechanism that is not yet fully defined.
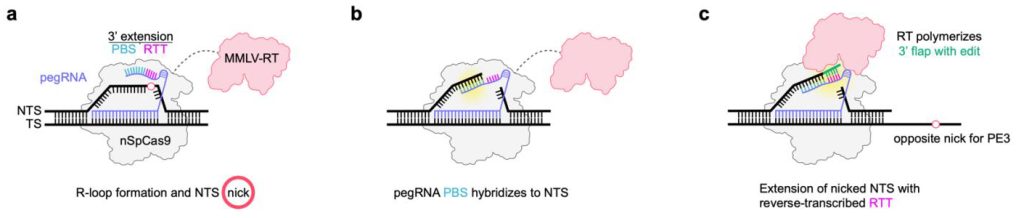
Optimizing prime editing
First authors Julian Grünewald, Bret R. Miller, and Regan N. Szalay from MGH found that the reverse transcriptases and nCas9 components of prime editing proteins function efficiently even when separate. The MGH researchers use this Split-PE (prime editing) system to rapidly identify and engineer more compact prime editor architectures that also broaden the types of reverse transcriptases used for prime editing.
The results strongly suggest that, with existing intact prime editing proteins, the reverse transcriptase activity is likely provided by a second prime editing molecule presumably not bound to the target DNA site, possibly from solution. This, in turn, implies that the efficiency of prime editing might be further increased by creating different next-generation fusions in which the reverse transcriptases actually must function in cis to the nCas9 (that is, a configuration in which reverse transcriptase activity is dependent on being tethered to the on-target site).
Improved Prime Editing Delivery
The Split-PEs and reduced-size reverse transcriptases described provide reagents and architectures that should enhance and better enable the delivery of prime editing components and accelerate further improvements to the platform. Split-PEs address a limitation imposed by size-constrained AAV vectors—that the full-length PE2 protein is too large to fit into a single AAV vector. By leveraging the Split-PE architecture, one can encode the nSpCas9 protein in one AAV and the pegRNA/gRNA and reverse transcriptase in another. This creates a configuration in which only cells transduced by both vectors will undergo editing without needing additional components.
The authors also note that our Split-PE system would be expected to enhance and simplify both RNA and ribonucleoprotein delivery methods due to more efficient expression of shorter-length nCas9 and reverse transcriptase components instead of a full-length fusion of these two components. This modularity should also permit the rapid screening of alternative nicking Cas9 or other nickases for prime editing.
Expanding the Prime Editing Toolkit
These studies demonstrate that the split architecture can facilitate more rapid screening of new prime editor variants with improved properties. Rather than cloning and sequencing a new lengthy fusion for each reverse transcriptase variant and determining where and how to fuse each of these to a nicking Cas9, the MGH researchers were able to construct rapidly and then screen an extensive series of different viral, non-viral, and engineered reverse transcriptases to identify those with desired activities.
This finding opens up the possibility of mining the vast number of non-viral, bacterial group II intron reverse transcriptases (G2I-RTs) that exist in diverse bacteria and archaea as starting points to engineer prime editors that are not only smaller but that may also provide improved function, such as higher fidelity or greater processivity. The enormous size of this treasure trove of reverse transcriptases that might be used for prime editing is evident from a recent computational search that yielded over 4,000 G2I-RTs.






