If your only CRISPR tool is an ordinary Cas9 enzyme, you might imagine that every genome editing problem has the same solution, a blunt double-strand break. Fortunately, ordinary Cas9 enzymes are being joined by more sophisticated CRISPR tools. The new additions to the CRISPR toolbox, which include modified Cas9 enzymes and non-Cas9 enzymes, are helping scientists accomplish familiar genome editing tasks more elegantly, as well as tackle new tasks beyond the reach of ordinary Cas9.
At present, the proliferation of new tools is sustaining and even intensifying CRISPR research, which is already robust. Over a thousand CRISPR-related articles have been published over the past year. Many of them highlight the various CRISPR effector modules and describe how they are being characterized and tested for nucleic acid editing, imaging, and disease diagnostics. Some of the new tools are opening up epigenomic applications select by tethering methyltransferases to enzymatically deactivated Cas9 proteins); others are enabling controllable activation of Cas9 select by inserting light-responsive domains into Cas9 proteins or engineering small-molecule-inducible guide RNAs); yet others are installing off switches into CRISPR systems select by adapting phage elements that degrade or inhibit Cas proteins).
Besides developing platforms that elaborate on Cas9 in various ways, researchers are exploring alternative Cas enzymes, such as Cas3, Cas12, and Cas13, as well as CRISPR-based transposon systems. The alternative platforms promise to bring about novel applications, provided that various platform-specific challenges, such as the targeted delivery of bulky CRISPR elements, can be overcome.
Heavy-duty tools
Many of the naturally occurring CRISPR-Cas effector complexes studied to date are capable of effectively targeting nucleic acids, but precise manipulation of genomes can be difficult. This limitation may soon be overcome, suggests Jennifer Doudna, PhD, a leader in the CRISPR revolution and a professor at the University of California, Berkeley. She cites recent research that has shown the potential of CRISPR-associated transposases to insert long pieces of DNA into locations along the Escherichia coli genome. “We will see,” she says, “if this approach is effective in eukaryotic cells, which would further expand the list of treatable genetic diseases.”
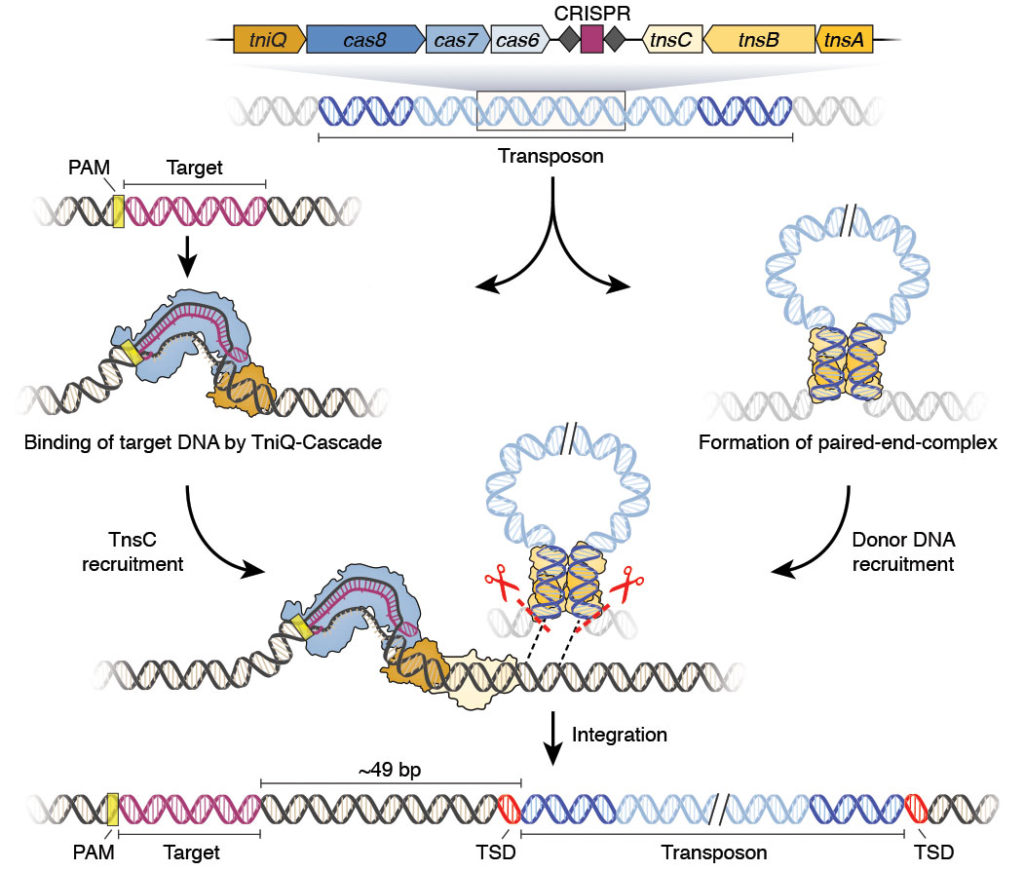
Also, researchers have shown the unique ability for the larger Type I CRISPR systems to introduce large deletions within a genome. Doudna suggests that the use of the Cas3 protein to unidirectionally remove long DNA segments provides an avenue to engineer minimal genomes, study regions of unknown function, and aid in metabolic engineering.
Another CRISPR-based approach may facilitate large-scale genomic manipulations. This approach combines a catalytically inactivated CRISPR effector protein with a transposon, and it can circumvent multiple issues that researchers commonly encounter when using traditional double-strand break genome engineering techniques.1
Although template-driven engineering via double-strand breaks has benefits, this approach can introduce unwanted insertions, deletions, or chromosomal translocations. But Doudna says that DNA integration via transposons does not cut the target sequence, which can potentially improve editing efficiency and work within nondividing cells.
“The use of transposons may also bypass the need for DNA templates with long homology arms, which can be difficult to fit into delivery vessels,” proposes Doudna. Another advantage of transposon-based CRISPR systems is cited by Kevin M. Esvelt, PhD, an assistant professor at the MIT Media Lab and leader of the lab’s Sculpting Evolution group. “The benefit of a CRISPR-based transposon,” he declares, “is it would let you add really large things.” Alternative techniques might be preferable if smaller insertions were desired, such as those compatible with in-frame editing.
As with most CRISPR tools, the latest CRISPR integrase systems have been given catchy acronyms. At the University of Columbia, the lab led by assistant professor of biochemistry and molecular biophysics Sam Sternberg, PhD, has introduced INTEGRATE select INsert Transposable Elements by Guide RNA-Assisted Targeting). This approach uses a Type I CRISPR system. At the Broad Institute, core institute member Feng Zhang, PhD, has led the effort to develop the CAST select CRISPR-associated transposon) method. It employs a Type V CRISPR system.
Imaging and diagnostic tools
There is a lot of excitement behind the CRISPR-based imaging and diagnostic tool called specific high-sensitivity enzymatic reporter unlocking select SHERLOCK). This tool, which was developed by Zhang’s group at the Broad Institute, relies on Cas12 and Cas13 and allows multiplexed, portable, and ultrasensitive detection of RNA or DNA from clinically relevant samples.2
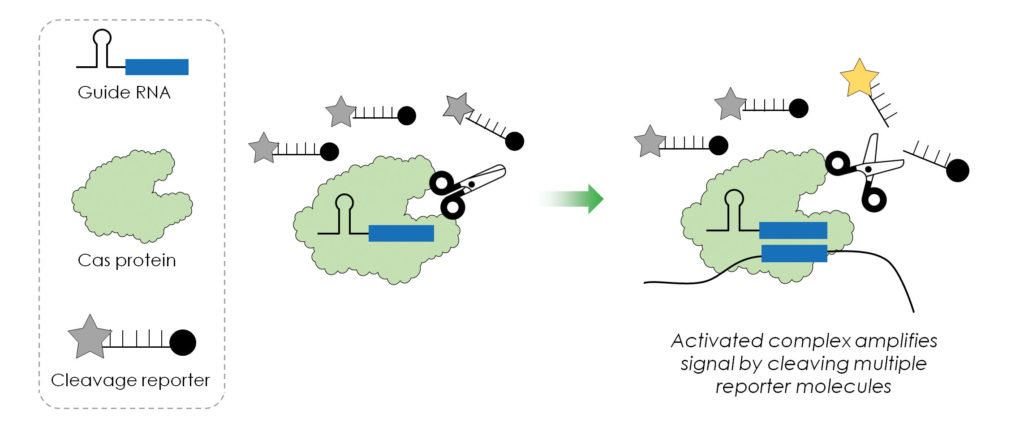
SHERLOCK excites Esvelt because he thinks it is by far the best diagnostic tool that we’re likely to see for some time. Esvelt says that SHERLOCK is particularly useful if you’re looking for a specific thing rather than just wanting to know what’s out there, where you’re pretty much forced to indiscriminate sequencing. “It’s basically single-molecule level detection for nucleic acids,” Esvelt elaborates. “For any given biological sample, you’re detecting a nucleic acid characteristic of a given agent, particularly if a pathogen is present at the fantastically low levels.”
Esvelt suggests that SHERLOCK can be combined with paper-based diagnostic technology, making it incredibly cheap, accessible, and employable. “That’s what we need for biosurveillance to prevent the next pandemic,” he emphasizes. “For known viruses, that’s what we want to be rolled out across the world. Certainly, on every transcontinental flight, ideally, everyone spits onto a piece of paper that then lights up if they’re infected with a particular virus.”
Another CRISPR-based diagnostic tool is DETECTR, which stands for DNA endonuclease targeted CRISPR trans reporter. Developed by Doudna’s lab and licensed from the University of California, Berkeley, DETECTR is currently being commercialized by Mammoth Biosciences. The tool, incorporates Cas12a/Cas13.3
Inducible and programmable tools
There is a lot of demand for better inducible CRISPR systems and, specifically, ones that would work in a mouse. “Right now, most mouse inducible knockouts still use inducible Cre recombinase,” Esvelt says. “It would be lovely if you could just do a single integration of a CRISPR system in the expression context of a given gene and tissue type—but inducible.”
This would allow researchers to just make one mouse and knockout one or multiple genes in combination depending on which guide RNA was programmed. Esvelt suggests that this could be controlled, for example, by adding a relevant small-molecule drug or shining a light on the mouse.
Precision tools
One of the most exciting advances in genome editing using CRISPR is that the ability to introduce specific base edits in the genome enables investigation of natural polymorphisms associated with human disease in a laboratory setting as well as potentially correct disease-causing polymorphisms in non-germline adult tissue in patients.
An alternative CRISPR-Cas9 gene editing system has been developed by a team of researchers from City University of Hong Kong and Karolinska Institutet. The system incorporates a modified Cas9 from Staphylococcus aureus and is called SaCas9-HF. Like other SaCas9 enyzmes, it is more compact than the Cas9 enzymes from Streptococcus pyogenes. It is, however, capable of greater precision.
Base editing is a CRISPR approach that eschews double-strand breaks and instead nicks a single strand of target DNA to convert one base pair to another. However, there have been problems. CRISPR-guided base editing has until recently been limited due to inefficient editing as well as unintended off-target effects.
A new approach called prime editing has been developed by the Broad Institute group led by David R. Liu, PhD, a core institute member and director of the Merkin Institute for Transformative Technologies in Healthcare. Prime editing has shown great promise in drastically improving efficiency and reducing off-target edits.4 Like base editing, prime editing also starts with nicking. But it has a different purpose. It allows users to delete, add, or rewrite small sequences to arbitrary new sequences.
In traditional editing, insertions follow the introduction of a double-strand break and require that a new template originate as a separate piece of DNA. In prime editing, an altered guide RNA is used as the template for the new sequence. It doesn’t require exogenous donor DNA. In Esvelt’s estimation, “It can become much more efficient to use prime editing at a site than to use traditional CRISPR editing.”
Prime editing may prove a valuable addition to the CRISPR toolbox used to study virus-host interactions, says Jan Carette, PhD, an associate professor of microbiology and immunology at Stanford University. “Designing a fundamentally different editing paradigm and clever engineering led to a system with much greater flexibility and efficiency,” Carette points out. He thinks that although it is early days, prime editing looks like a breakthrough technology with near-limitless possibilities in biomedical research.
To unlock the full potential of prime editing, several technical hurdles need to be overcome in scaling up the technology. For example, prime editing in a large pooled format would allow for the creation of large cell libraries representing virtually all pathogenic genetic polymorphisms. Carette is convinced that, given the rapid pace of development in the CRISPR field, that this problem will be solved: “The ability to generate the full genetic diversity in a tissue culture dish will make the goal of precision medicine based on individualized therapies more attainable in cancer therapy and infectious disease.”
Getting tools to the worksite
Doudna says that the delivery challenge—how we decide to carry and transport CRISPR tools—is an area that requires cross-disciplinary collaboration and ingenuity. Current genome editing treatments for genetic diseases rely on an ex vivo approach, which involves removing cells from a patient and performing the gene edit before reintroducing the engineered cells.
Recently, ex vivo gene modification has been used to treat blood disorders. For example, a CRISPR-Cas9 gene editing therapy called CTX001 is being evaluated for its ability to boost fetal hemoglobin in stem cells. This is encouraging news, even though CTX001’s developers, CRISPR Therapeutics and Vertex Pharmaceuticals, have released only preliminary data from their Phase I/II trial.5 “This could be transformative,” observes Carette, “and it would be critical to see at what price point this therapy will be offered.”
“If researchers could effectively deliver CRISPR complexes directly into the body and target specific cell types,” declares Doudna, “we could unlock the potential to treat a much wider range of genetic diseases. “In 2020, I think we will witness a surge in exciting in vivo delivery approaches making use of nanoparticles, lipid vesicles, RNP complexes, viruses, and more!”
The decade ahead
Scientists have only begun to understand and harness the potential of CRISPR-Cas systems across bacteria, archaea, and even phages, targeting and manipulating RNA and DNA. As scientists continue to combine genomics, biochemical characterization, and structural analyses, we are bound to uncover new CRISPR systems in the decade ahead.
“Within 5 to 10 years,” predicts Doudna, “scientists will be using [transposon-based CRISPR] systems along with Cas9, base editing, prime editing, and tools yet to be created, to introduce any change, at any genetic location, in any organism.”
References
1. LeMieux J. CRISPR Jumps in a New Direction. GEN. June 12, 2019.
2. CRISPR Diagnostics. GEN. May 11, 2018.
3. Mullin E. CRISPR Dx May Help Nix Disease. GEN. October 1, 2019.
4. LeMieux J. Genome Editing Heads to Primetime. GEN. October 21, 2019.
5. CRISPR Therapeutics, Vertex Report First Data from Trials of Gene-Editing Treatment CTX001. GEN. November 19, 2019.
Liu Lab Targets Off-Target Editing
The Harvard University lab led by David R. Liu, PhD, introduced a pair of genome editing systems—a cytosine-to-thymine base editor select CBE) and an adenosine-to-guanosine base editor select ABE). At the center of both systems is a Cas9 protein that nicks DNA instead of creating a double-strand break. In the CBE, the Cas9 “nickase” is fused to a cytidine deaminase; in the ABE, an adenosine deaminase. select In the CBE system, a uracil glycosylase inhibitor, interferes with a repair mechanism that would restore cytosine.)

David Liu, Ph
Base editing systems make point mutations—something that generic CRISPR-Cas systems cannot easily do. Although this new ability makes base editing systems welcome additions to the CRISPR toolbox, they must be used with caution. Besides making point mutations, they produce low-level, widespread, hard-to-find, off-target DNA edits.
“This type of off-target editing can occur at random locations in the genome,” notes Liu. “When you run the experiment 10 times, you get 10 different sets of answers.” This variability, he continues, “makes it particularly challenging to study Cas9-independent off-target editing.”
It’s unclear if this type of off-target activity will result in adverse effects. Nonetheless, Liu maintains, his group has “a responsibility as part of the gene editing community to try to understand and minimize all types of unwanted editing.” So, the group is working to detect off-target activity without requiring expensive and slow whole-genome sequencing.
Last month, a Liu lab team led by graduate students Jordan Doman and Aditya Raguram published a study in Nature Biotechnology that describes multiple, rapid, inexpensive assays to detect off-target editing in both bacterial and mammalian cells. Using these assays, the lab developed “a suite of next-generation CBEs” that retain on-target editing activity while exhibiting greatly reduced Cas9-independent off-target DNA and RNA editing select ~10–100-fold lower off-target editing, similar to untreated control cells).
Liu recommends that researchers begin using these new variants for experiments in which off-target editing is “a major concern.” He asserts that the variants will be particularly useful for therapeutic applications where off-target editing must be minimized.


