A reasonable idea, suggests Sekar Kathiresan, MD, co-founder and CEO of Verve Therapeutics, is the development of a one-time treatment—a single spelling change in the DNA of a liver gene in an adult person—that would “turn off the gene and lower blood cholesterol levels for the rest of the person’s life.” Although this idea would not have been taken very seriously 10 years ago, it is now being investigated. It is a potent illustration of the magnificent promise of genome editing in general, and of base editing in particular.
Millions of people manage heart disease with daily doses of cholesterol-lowering drugs such as statins. But a one-and-done approach is on the horizon thanks to base editing, a budding technology in the genome editing space. Since its first publication in Nature and Science in 2016, base editing has made rapid strides through preclinical studies and is poised to revolutionize medicine.
This precise technology holds the key for a new class of genetic medicines developed through direct correction of disease-causing mutations and other genetic manipulations—inserting protective genetic variants, activating or silencing regulatory elements, knocking out genes through stop codon or splice site mutations, and potentially realizing a multiplexed approach.
Beam them out, Scotty
Transitions within the two base groups (purines and pyrimidines) responsible for diseases such as sickle-cell disease, Tay-Sachs disease, and several types of cancers collectively account for more than 60% of human pathogenic point mutations.
CRISPR, although efficient at knocking out genes, can typically introduce single base changes only at rates of 0.1% to 5%. In addition, it risks introducing many random insertions and deletions (indels) through zealous cellular repair systems that patch over CRISPR-generated double-strand breaks.
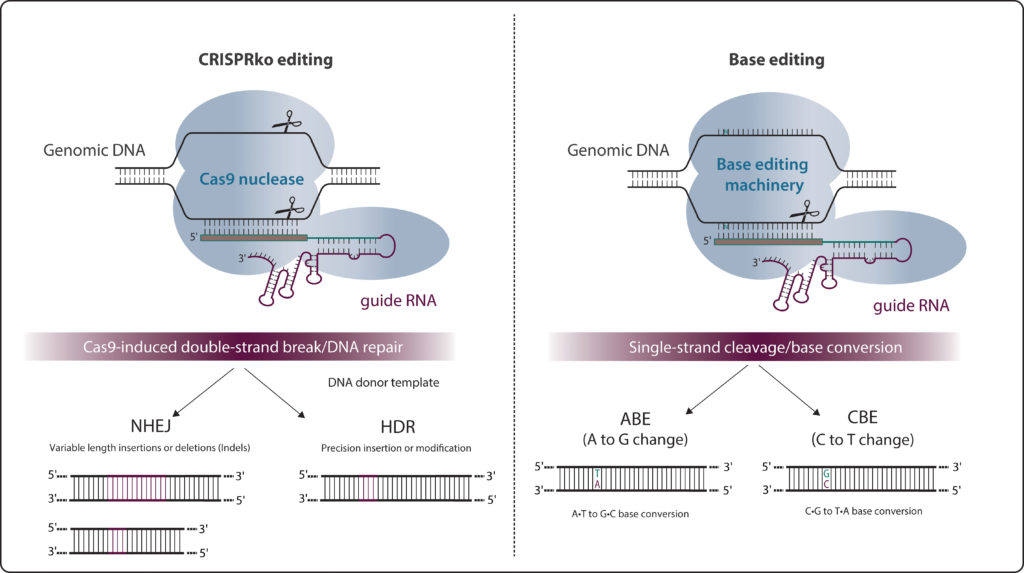
As detailed in a 2019 GEN article (“All About That Base Editing”), base editors consist of a partially inactive Cas9 nickase (nCas9) connected to a deaminase. This complex drives specific, accurate, and permanent single nucleotide changes in the genome without introducing double-stranded DNA breaks. The first base editors—cytosine base editors (CTEs) that made it possible to change a cytosine to a thymine—were independently developed in 2016 by two groups, one led by David Liu, PhD, at Harvard University, and one led by Akihiko Kondo, PhD, at Kobe University.
Liu co-founded Beam Therapeutics with colleagues Feng Zhang, PhD, and Keith Joung, MD, to expand the use of base editing to develop precision genetic medicines. Bio Palette, a Japanese biotech company that has licensed base editing intellectual property from Kobe University, has partnered with Beam Therapeutics to develop human therapeutics.
Barely five years from the development of base editing technology, Liu and a variety of collaborators have produced some powerful preclinical findings. In a breakthrough study published in Nature, the Liu laboratory, working with NIH director Francis Collins, MD, PhD, used an adenine base editor (ABE) to durably correct a single base in both alleles of the LMNA gene in a mouse model of progeria, a rare genetic disease where the buildup of a toxic protein induces premature aging and death.
“Fixing the tiny DNA typo responsible for progeria is now within our grasp and getting closer to landing in the ‘can do’ category,” Collins wrote on the NIH Director’s Blog. (For more information, see “Base Editing Shows Promise for Progeria in Mouse Model,” a news story that was posted to the GEN website January 6, 2021.)
In recent months, genome editing has seen a surge not just as a promising medical technology but also in the stock market. Brad Loncar, CEO of Loncar Investments, in a GEN Edge webinar (“Trends and Takeaways from J.P. Morgan 2021”) that aired January 27, 2021, expressed optimism that CRISPR will change medicine in the coming decade, but worried that some stocks had become overvalued. For example, he remarked that Beam “is a company that is a year from the clinic and has a $6-billion valuation.” And he noted that some leading CRISPR companies have overlaps in their pipelines.
“Currently, Beam Therapeutics is the only company that holds a base editing license for therapeutics,” says Nicole Gaudelli, PhD, who developed ABEs when she was a postdoc in Liu’s laboratory. She is currently head of gene editing technologies at Beam. Along with several other companies targeting sickle-cell disease, Beam has two drug candidates for hemoglobinopathies in its pipeline.
“Beam-101 uses an adenine base editor for sickle-cell anemia and upregulates gamma globin by disrupting the promoter that drives it,” Gaudelli asserts. “The other approach that I’m more excited about—and that highlights the power of base editing—replaces the sickle-cell disease–causing point mutation with a nonpathogenic variant.”
Editing out blindness
Leber congenital amaurosis (LCA) is a group of recessively inherited retinal disorders caused by loss-of-function mutations in several genes that are important for retinal function. Patients start to lose vision in childhood and are legally blind by 20 or 30. At present, there is no cure for LCA. Luxturna, the FDA-approved gene therapy for one type of LCA, targets patients who have mutations in both alleles of the RPE65 gene.

“Gene augmentation improved visual sensitivity in the patient during the first year after surgery, but after a year, patients showed retinal degeneration and decline of visual function,” says Susie Suh, an MD-PhD student at Case Western Reserve University. “If we can correct the mutation endogenously in a precise manner, that could be another therapeutic modality.”
Working in the lab of Krzysztof Palczewski, PhD, at University of California, Irvine, Suh first used conventional CRISPR-Cas9 editing to restore a functional RPE65, a gene that is exclusively expressed in a single layer of cells behind the retina. But the method caused more indel mutations than desired. She then tried base editing using a lentivirus-packaged ABE transfected in cells, although her target did not contain a protospacer adjacent motif (PAM)—a requirement for base editing.
“I didn’t think I would have a rescue of RPE65, but a Western blot showed RPE65 expression restored in base editor–transfected cells,” Suh relates. Suh and colleagues went on to treat live mice and saw a restoration of RPE65 gene function, up to 29%, with minimal indels or off-target mutations—sufficient to restore vision in mice. Suh’s team collaborated with David Liu’s group to demonstrate the lack of off-targeting effects in mice; the work was recently published in Nature Biomedical Engineering.
Base editing “can address both recessive and autosomal dominant inherited dystrophies,” says Suh. To move this approach into the clinic, the team is testing the durability of the base correction, checking for off-target effects, and switching to an adeno-associated virus (AAV) delivery system.
Commenting on the LCA study in a recent interview with Human Gene Therapy, Liu said, “Even moderate levels of editing can show substantial rescue. [The] relationship between extensive editing and the amount of rescue of the disease phenotype is one of the most compelling reasons to be optimistic about the use of gene editing to treat genetic disease.”
Attacking heart disease
Base editing’s potential lies beyond treating rare Mendelian diseases. Verve Therapeutics is set to use base editing in developing drugs for coronary heart disease, the world’s leading cause of heart attack and death.

Verve CEO Sekar Kathiresan, MD, says, “Some people are protected naturally from heart disease by mutations in any of eight different genes that turn off a cholesterol synthesizing gene in the liver.” Individuals harboring protective mutations in PCSK9 and ANGPTL3 have low levels of cholesterol and are resistant to heart attacks. Kathiresan plans to “develop a gene editing medicine that would mimic those natural resistance mutations.” Since launching Verve in 2018, his team has spent two years performing experiments in cells, mice, and monkeys to develop this drug.
Verve’s lead product, Verve-101, will be tested initially in individuals with familial hypercholesterolemia. Early data show that a one-time treatment is durable for six months or more in primates, Kathiresan reports. Verve-101 consists of a base editor that makes a gene-inactivating adenine-to-guanine substitution in PCSK9 (thereby lowering cholesterol) and a guide RNA to target the enzyme complex. These components are packaged in a lipid nanoparticle delivered to the liver.
“In monkeys, a one-time infusion turned off PCSK9,” Kathiresan details. Two weeks after injecting Verve-101, levels of PCSK9 production decreased by 90% and LDL cholesterol levels dropped by 60%. After six months, “we get the exact same levels of reduction,” Kathiresan adds. “We expect this to be durable for the lifetime of the animal.”
The advantage of targeting PCSK9 is that Verve-101 should be effective in treating patients who have elevated cholesterol levels due to other mutations, such as in apolipoprotein B-100 or LDLRAP1, or even due to lifestyle-related reasons. This was explicitly part of Verve’s development strategy.
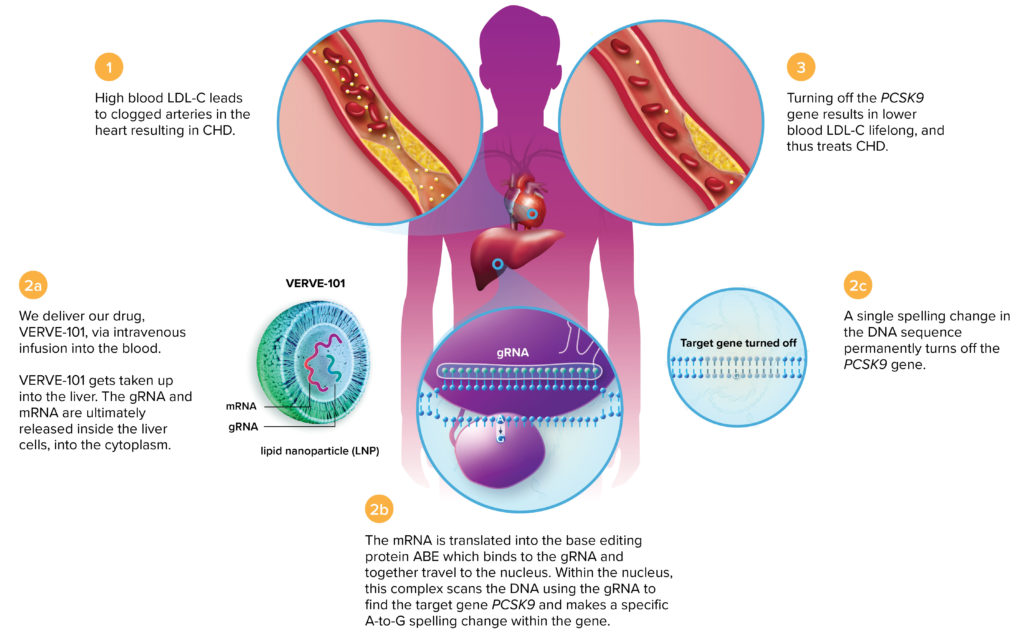
gene. Having achieved long-term reductions in LDL cholesterol levels in cells and animal
models, Verve-101 is poised to enter human trials.
“We think of our clinical development as a bulls-eye model,” Kathiresan declares. “The center of the bull’s eye is the genetic form of the disease. That is about 1.6 million patients in the United States and Europe. The next ring of the bull’s eye is anybody with atherosclerotic cardiovascular disease. That is about 24 million patients in the United States and Europe. And the next tier is true prevention, which is an even larger population.”
Therapeutic edits for cancer
Delivery is a major area of investigation in base editing. Base editors are larger than the standard CRISPR-Cas9 machinery, requiring more effort to synthesize mRNA and functional protein, and different AAV delivery strategies, such as using a two-part packaging system, or a smaller inactivated nuclease.
“We are focusing more on the licensing and the technological development than on product development,” says Jennifer Harbottle, PhD, senior scientist in the R&D Base Editing team of PerkinElmer’s Horizon Discovery business. “On the application side, we’ve chosen to focus our energies on next-generation cell therapies because you can manipulate the cells ex vivo.”
Modifying cells outside the body and transplanting them back into patients as therapeutic agents simplifies the standardization of safe and effective delivery strategies. The company’s base editing technology is licensed from a Rutgers University group led by postdoctoral fellow Juan Carlos Collantes, PhD, and principal investigator Shengkan Jin, PhD. Harbottle co-authored a research article in the February 2020 issue of The CRISPR Journal on the development of the Rutgers-Horizon base editing platform that the company is using to develop immune-based cell therapies, particularly chimeric antigen receptor (CAR) T-cell therapies to treat various types of cancers.
Harbottle and her team are using base editors to knock out key checkpoint inhibitors in engineered T cells that would increase their persistence and efficacy in the tumor environment. “This is where the beauty of base editing comes into play, Harbottle explains. “We’re able to knock out lots of genes at once without generating off-target effects or at least without generating indels.”
Beam Therapeutics is developing BEAM-201, a CD7-targeting CAR T cell for the treatment of T-cell acute lymphoblastic leukemia. “It uses a cytosine base editor to generate modified CAR T cells using multiplex base editing,” Gaudelli says.
Raising hopes, clearing hurdles
Computing support is key for the rapid progress of a data-intense field such as base editing. Benchling, a company devoted to powering R&D in the life sciences, has developed a suite of cloud applications to design guide RNAs for base editing. These applications incorporate complex scoring algorithms to score the guide RNAs in terms of their on-target efficiency and potential off-target binding.
“Back in 2016 when the initial base editing paper came out, we worked with the scientists from David Liu’s laboratory to add guide design capabilities for base editing to our existing CRISPR designing tools,” says Ashu Singhal, co-founder and president at Benchling. The user interface also helps scientists collaborate on experiments and manage research workflows.
Delivery is paramount for the best function of a base editor, especially in therapeutic applications. “And for that you need mRNA or ribonucleoprotein (RNP) delivery that is transient and more effective,” stresses Harbottle. Plasmid delivery leads to the prolonged retention of the base editing machinery in the living system, increasing the likelihood for undesired mutations in the genomic DNA or deamination in mRNA. Consequently, this form of delivery is not suitable for clinical therapeutics.
Despite the expanding catalog, most base editors developed so far need to be delivered by plasmids. Few variants have been optimized for mRNA or RNP delivery systems. “That’s where it ends up being a lot more limited than the data that we have for any of the standard or special base editors,” Harbottle points out.
“The technology works great in the Petri dish,” says Gaudelli, “but getting it to the tissue is the challenge.”
The presence of genetic variations in the editing window in different populations might result in the generation of unintended bystander mutations depending on the sequence context, the deaminase, and other prerequisites for optimized base editing.
“The key is to find a site in the gene that is generalizable in the population and does not have a lot of genetic variation,” specifies Kathiresan, who is applying base editing primarily to inactivate genes by introducing stop codons. For other base editing strategies, it might be more difficult to dictate the genomic sequence around the target base.
Frontier research on base editing is focusing on developing different types of base editors with specific and often diametrically opposite attributes. “Some base editors are focused on being as precise and pure as possible, where you just want to change a single nucleotide of interest without having any effects elsewhere,” Harbottle states. “These are tailored for clinical applications downstream. [But some laboratories] are harnessing the system to generate a hyperactive base editor that allows trait diversification that help in developing, for instance, in vitro drug discovery platforms.”
Additions to the genome editing toolbox include increasingly refined tools. The latest of these is prime editing, which was developed in Liu’s group by Andrew Anzalone, MD, PhD, and first published in Nature in 2019. Using a prime editing guide RNA (which identifies the target site) and a catalytically inactive Cas9 fused to reverse transcriptase, prime editing can introduce small insertions and deletions in addition to a full repertoire of single-base changes. Liu founded a new company, Prime Medicine, to develop the technology. It’s still early days, but prime editing could correct disease mutations that other tools in the genome editing toolbox cannot.