Sponsored content brought to you by

If you have ever screened a compound library and successfully killed tumor cells in a dish, you might be surprised to find that many of those compounds may have little or no effect when those same tumor cells are implanted in a mouse. Certainly, failing your lead compound sooner, rather than later, in the drug development pipeline will save you time and resources. This idea is especially relevant when taking your lead compound from the preclinical space and transitioning into the clinic.
At JAX, our In Vivo Pharmacology Services group has run over 50 efficacy studies using the JAX Onco-Hu™ Immuno-oncology platform to test novel immune checkpoint inhibitors and other novel immunotherapies targeted at PD1, PDL1, CTLA4, OX40 and IDO1, for example. To us, a successful study is one in which the client has scientifically sound data in hand and is able to quickly determine whether their compounds should continue along in the drug pipeline or be repurposed.
I’ve gathered a few tips for generating robust data using Onco-Hu™ mice in immuno-oncology drug efficacy studies. Many of these tips could be applied to other mouse oncology models.
Pro Tip 1: Know thy mouse and pick the “best” one(s) to do the job
It can be tempting to call our Technical Information Services (TIS) group at JAX and ask us to select the “best” mouse for immuno-oncology research. Realistically, there isn’t just one best model. Each mouse model serves a unique purpose, so you need to know whether the model you select will answer the question you are asking.
For example, the JAX In Vivo Pharmacology services group routinely co-engrafts CD34+ hematopoietic stem cells (HSC) from cord blood and either Patient Derived Xenografts (PDX) or human cancer cell lines into the severely immunodeficient NSG™ (005557) or NSG™-SGM3 (013062) mouse strains, creating what we call Onco-Hu™ mice. Amazingly, many cancers grow in the presence of an HLA mismatched immune system, likely through an array of immune evasive mechanisms. We have demonstrated efficacy in this platform using standard of care (SoC) therapeutics such as Keytruda and Yervoy.
Based on this model, you can query several aspects of tumor biology. The Onco-Hu™ model can help determine
- whether new immune checkpoint inhibitors can reinvigorate exhausted T cells
- which tumor types will be most responsive to immunotherapy
- optimal therapy combinations
- mechanisms of tumor immune cell evasion and resistance to immunomodulatory agents
The mechanism of action (MOA) of your lead compound and your position in the drug development pipeline should be considered to determine whether the Onco-Hu™ model is appropriate for your study. Alternatively, because it is a pretty sophisticated model, the model may not have been validated for a particular MOA yet. We’re continually learning more about the Onco-Hu™ model capabilities, especially as we employ other NSG™ variant mice to further promote development of other human immune cell populations in more advanced models.
In the spirit of knowing more about your model selection, download our guide “Comprehensive Evaluation of Human Immune System Reconstitution in NSG™ and NSG™-SGM3 Mouse Models” to learn what immune cell types are present.
Pro Tip 2: Expect variability in PDX response and use it to your advantage
In the Onco-Hu™ model there are 2 major sources of variability: PDX donor and the CD34+ donor. Both of these components contribute to the variability of the response to your therapeutic, and reflects what is observed in the clinical setting.
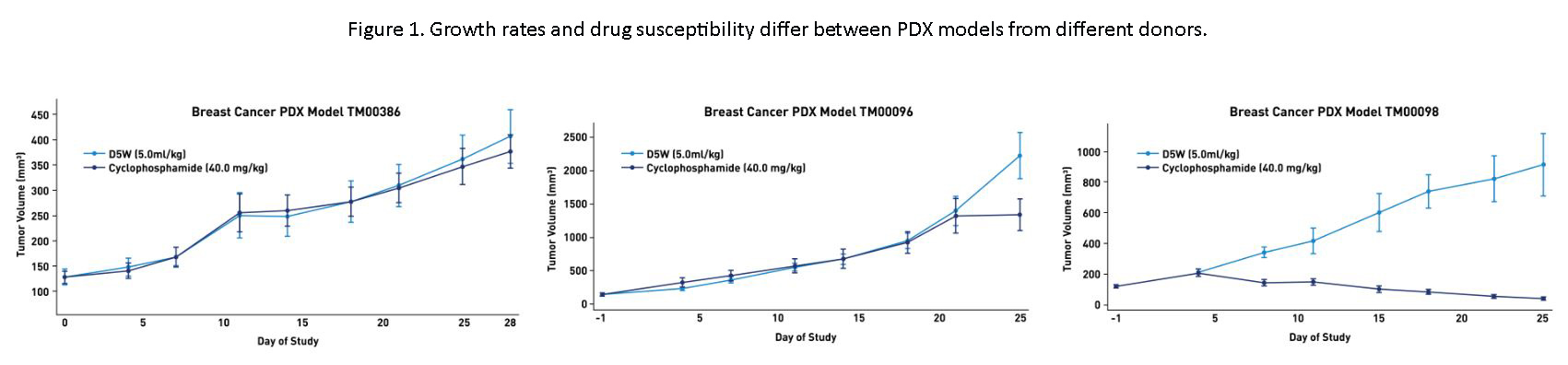
From the PDX standpoint, unlike syngeneic and cell line xenograft models, PDX models are heterogeneous. Heterogeneity exists between PDX models, such that they differ in growth rate and drug susceptibility (Figure 1); this more accurately reflects patient-to-patient variability that does not exist in homogeneous cell line models. Experimentally, using more than one PDX sample may more accurately predict whether your compound will have efficacy in a variety of patients of one cancer type.

Heterogeneity also exists within a single PDX model, such that each mouse engrafted with the same PDX may differ in growth rate and drug susceptibility (Figure 2); this more accurately reflects variability of response of the multiple cell types present in a single tumor or multiple metastases within a single patient. Experimentally, this means you might need to run a pilot study to calculate the sample size necessary to power your results appropriately.
Pro Tip 3: Expect variability in CD34+ response and use it to your advantage
From the standpoint of CD34+ donors, we have found that some CD34+ donors respond well to the PD1 inhibitor, Keytruda (which we use as our SoC), and some do not (roughly 1/3). For those CD34+ donors who do not respond, we have confirmed that Keytruda has bound to its intended target within the tumor, so it is working the way we would expect. In these cases, we determined that these specific CD34+ donors do not respond to Keytruda, similar to what occurs in the clinic.
If you use only one CD34+ donor for the generation of experimental mice and it happens to not respond to the SoC, you may lose an entire experiment due to poor planning. You won’t know if absence of response of your compound is a failure of the compound or is because the CD34+ donor sample just does not respond to your compound. We recommend using 2-4 different CD34+ donors distributed evenly throughout control and experimental groups (with sufficient sample size) to protect the experiment from being lost.
The heterogeneity of Onco-Hu™ models, while challenging to work with experimentally, offer a more accurate reflection of the heterogeneity inherent in patient populations. When using Onco-Hu™, you will be in a better place to understand potential variability of response when your drug enters the clinic. For those donor samples that do not respond to your therapy, you can take the opportunity to collect samples that may help to understand mechanisms of resistance. Perhaps combination strategies using alternative checkpoint inhibitors will point the way. Maybe not.
Pro Tip 4: Be careful, level of T cell infiltration isn’t always a good predictor of response
Tumors can be described as “hot” or “cold,” indicating relative level of T cell infiltration as measured by either FACS or immunofluorescence staining. It can be tempting to think that “hot” tumors with high T cell infiltrate will have an equally strong reduction in tumor volume. Unfortunately, we have found this not to be true. For instance, sarcoma model TM00381 has a very high level of T cell infiltration, but we have yet to find a checkpoint inhibitor that has efficacy in this model. Likewise, breast tumor model TM00098 has a relatively low level of T cell infiltration, but responds well to Keytruda time and time again. Amount of T cell infiltration seems to be a function of the tumor itself and not of the efficaciousness of response.
As you might expect, this blog isn’t a definitive guide about how to design fail-proof immuno-oncology studies. Do you have more questions about immuno-oncology or have an experiment in mind that we might be able to help with? Drop us a line and let us know what you would like to test. We’ll see if we have the right model to push your drug discovery further.