Drug target identification has a key role in the drug discovery value chain. A critical step in the development of pharmaceuticals is identifying the direct targets of potential drug candidates as well as distinguishing any secondary or off-target effects.
One method of drug target identification, phenotypic screening, involves the addition of compounds to cells (or small model organisms) and measuring the impact on the phenotype or cell activity of interest. For compounds with a desirable impact on phenotype or cell function, the genes or gene products (that is, the targets) that the active compound directly perturbs must be identified. Thus, a critical step in the development of pharmaceutical drugs is identifying the direct targets of the active compounds, as well as any secondary or off-target effects of that compound that may impact further development.
In recent years, it has been established that mitochondrial and cellular metabolic processes are central to cell differentiation, cell proliferation, immune cell responses, hypoxia sensing, and apoptosis, in addition to their well-known roles of substrate oxidation and ATP production. Indeed, mitochondrial and metabolic dysfunction has increasingly been linked to a multitude of pathologies including cancer, immune cell and system disorders, neurodegeneration, cardiac disease, obesity and diabetes, and the aging process.
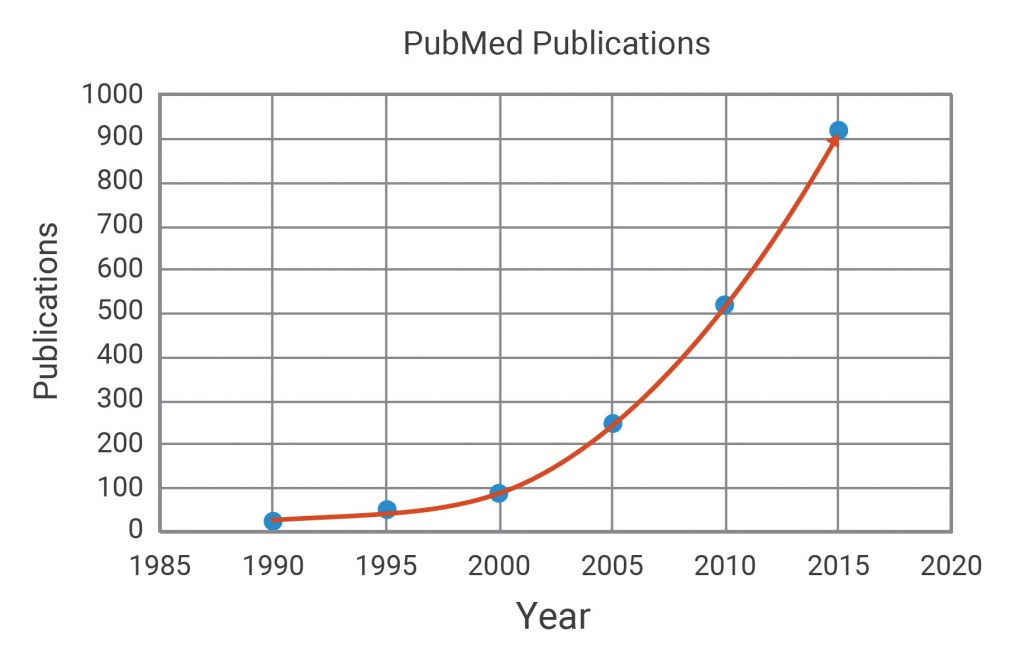
As such, interest in mitochondrial and metabolic drug targets has increased dramatically (Figure 1). Thus, there is a corresponding need for sensitive and direct measurements of metabolic pathway function to elucidate the specific (and any possible nonspecific) targets of potential drug candidates.
The Agilent Seahorse XFe96 Analyzer directly measures mitochondrial respiration and cell metabolism in live cells in a multiplate format. It is an ideal system for examining the functional effects of drugs targeted to mitochondrial and other metabolic pathways, such as glycolysis. This tutorial provides a general overview of the Seahorse XF applications and workflows that can be applied to metabolic target identification studies.
Identifying mitochondrial and metabolic drug targets
The Seahorse XF workflow for identification of mitochondrial and metabolic drug targets is divided into a series of assays designed to answer these main questions:
- Does the compound affect mitochondrial or metabolic function?
- What is the specific target of the compound?
- Are there any nonspecific or off-target effects?
For compounds (for example, drug X) that exhibit an effect in a phenotypic screen, an Agilent Seahorse XF Cell Mito Stress Test (MST) is performed to determine whether the compound affects mitochondrial function. This assay tests several key parameters of mitochondrial respiration as measured by oxygen consumption rate (Figure 2, left panel). Information about parameter changes (as well as the magnitudes of any changes) may indicate whether the compound is altering mitochondrial function. The results of this assay can also determine which types of follow-up XF assay designs are best suited for gathering more specific information, including drug target identification. As an example for drug X, the workflow will be applied to the well-known mitochondrial pyruvate carrier inhibitor, UK5099.
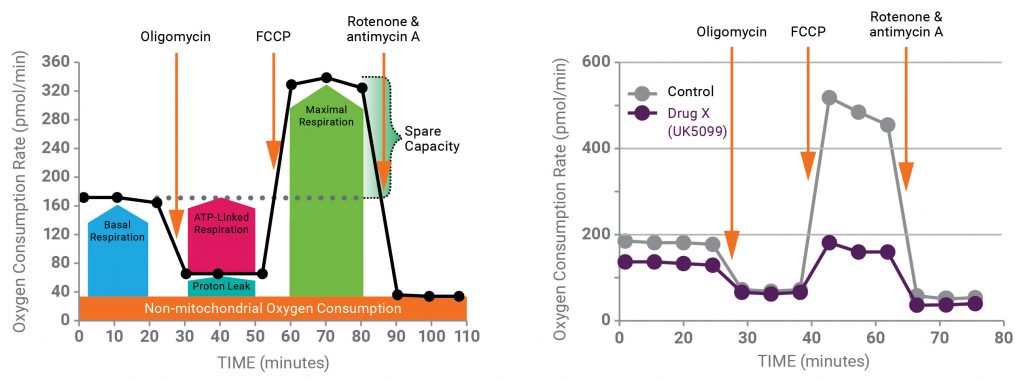
Figure 2 (right panel) shows the results of the MST run in the absence and presence of UK5099. The data demonstrate that UK5099 does indeed affect mitochondrial function, as evidenced by decreases in both basal and maximal respiration rates.
Next, which components of metabolism might be driving this change must be considered. The MST profile of UK5099 suggests that dysfunction occurs in substrate oxidation and/or the electron transport chain/oxidative phosphorylation pathways. These pathways include substrate transport and activities of rate-controlling proteins and enzymes, including glutaminases, CPT1a, pyruvate dehydrogenase (PDH), tricarboxylic acid (TCA) cycle enzymes, and electron transport and oxidative phosphorylation machinery.
To localize the effect of UK5099, Agilent Seahorse XF Plasma Membrane Permeabilizer (PMP) is used. Permeabilization of the plasma membrane allows direct access to the mitochondria with respect to substrate provision without physically separating the mitochondria from the cells. Because different oxidizable substrates feed into different metabolic pathways, permeabilized cells offered specific substrates may present respiration rates that may be used to identify the target that was modulated to cause the changes in mitochondrial respiration observed in intact cells. The substrate-dependent pathways for pyruvate, glutamate, and succinate are outlined simply in Figure 3.
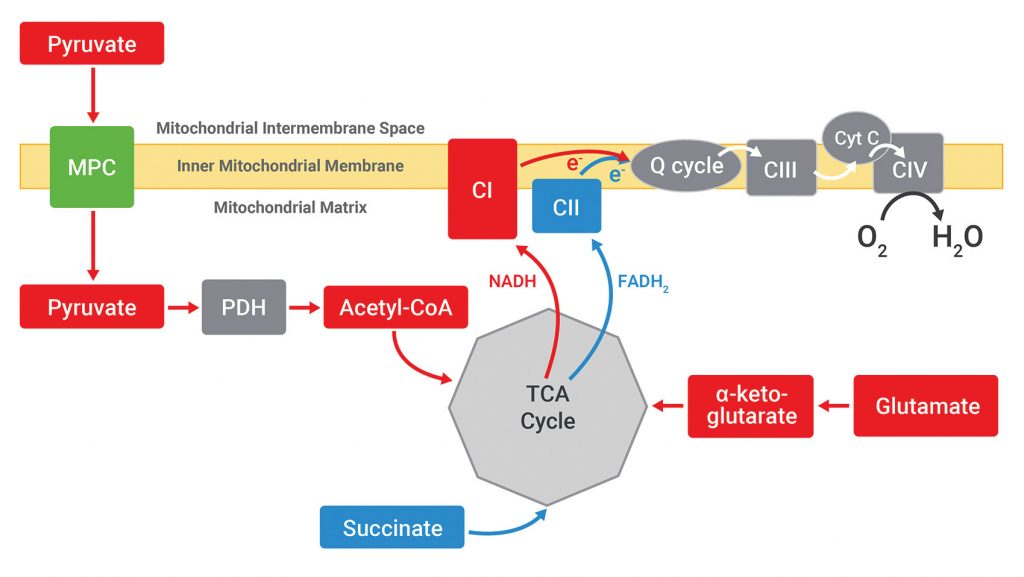
Thus, the next XF assay in the workflow is to provide these three substrates individually to permeabilized cells in the presence or absence of the drug candidate, UK5099. As shown in Figure 4, UK5099 blocks respiration only when pyruvate is the substrate; there is no effect when glutamate or succinate is provided to each type of permeabilized cell (human skeletal muscle myocytes, neonatal rat ventricular myocytes, and primary cortical rat neurons).1
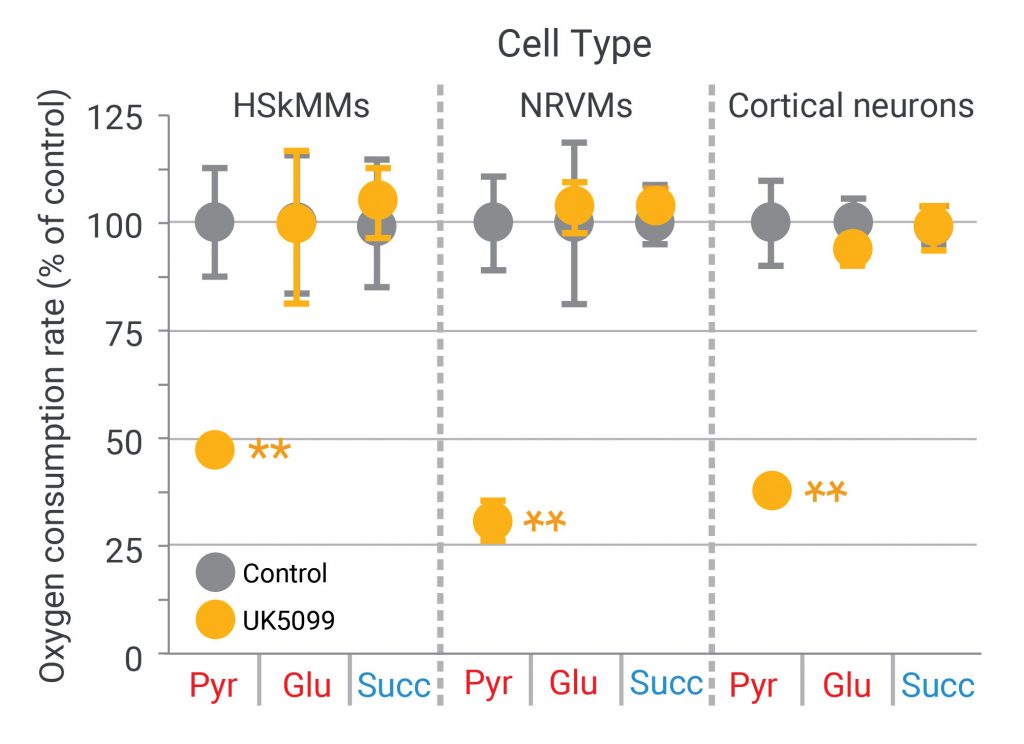
(Adapted from Divakaruni AS, et al. Thiazolidinediones are acute, specific inhibitors of the mitochondrial pyruvate carrier. PNAS 2013; 110(14): 5422–5427.)
Taken together, these results indicate that neither Complex I nor Complex II is the target of UK5099, and that the inhibition of respiration by UK5099 must lie upstream of both Complex I and the TCA cycle, since neither glutamate (Complex I substrate) or succinate (TCA/Complex II substrate) oxidation is affected. Moreover, these results also suggest that pyruvate dehydrogenase (PDH) or the mitochondrial pyruvate carrier (MPC) may be the target of UK5099.
Further assays with permeabilized cells and alternative substrates can then be performed to distinguish between PDH and MPC, as was done to demonstrate that the MPC is the specific target of UK5099.1
Summary
Our understanding of metabolism has evolved from seeing it perform simple housekeeping functions to seeing it play a central role in many normal and disease states. Examining the impact of compounds on mitochondrial function and metabolic phenotype in the context of live cells provides an avenue into the identification of targets for metabolic modulation. This approach complements other methods such as targeting signaling pathways and cell receptors.
The example outlined here highlights the importance of considering multiple mitochondrial pathways, including substrate transport and mitochondrial enzyme activity, in addition to electron transport and oxidative phosphorylation components. By incorporating direct cell-based measurements of mitochondrial and metabolic function into drug target identification studies, valuable and important insights regarding specific and nonspecific effects of compounds may be obtained.
George W. Rogers is R&D scientist and expert (Seahorse Products and Applications), Sarah E. Burroughs is product manager (Seahorse XF Instruments), and Brian P. Dranka is head of application development (Cell Analysis Division) at Agilent Technologies. The authors wish to thank Ajit Divakaruni for his research (as highlighted in “Thiazolidinediones are acute, specific inhibitors of the mitochondrial pyruvate carrier”) and acknowledge the influence his manuscript had on this tutorial.
Reference
1. Divakaruni AS, et al. Thiazolidinediones are acute, specific inhibitors of the mitochondrial pyruvate carrier. PNAS 2013; 110(14): 5422–5427.