The Biopharma 4.0 paradigm has a grand vision for the factory of the future. It envisions a factory where “smart” processes continuously learn from data to predict optimal states and control themselves to stay within this optimal space. The smart processes will also communicate with each other to control plant-wide logistics and supply chains to minimize costs and maximize productivity.
This paradigm shift in biomanufacturing will exponentially improve the flexibility and robustness of processes to result in safer, better, and more affordable biologics. At the heart of this paradigm shift is a digital transformation of biomanufacturing processes. The development of Digital Twins is a vital step towards this transformation.
Digital Twins are virtual counterparts of physical systems or processes. They enable predictive manufacturing. For example, Digital Twins can analyze, optimize, predict, and control physical processes in real time. They convert the physical process to a smart process and thus achieve the ultimate goal of the digital transformation.
This article focuses on three themes: how Digital Twins can revolutionize cell culture process development, how Insilico Biotechnology’s Digital Twins (DTs) bring together mechanistic modeling and artificial intelligence (AI), and how Digital Twins can be applied to realize the factory of the future.
There are two main factors driving the digital transformation of cell culture processes. On one side, advances in miniaturization and high-throughput data from cell culture processes demand superior data analytics methods. On the other side, the transformation of the biopharma industry toward personalized medicine demands higher flexibility, efficiency, and robustness from manufacturing processes. In particular, the development of cell culture processes, mainly carried out through ad hoc experimental approaches, is in dire need of an overhaul because of the inordinate impact of cell culture processes on product quality and cost of goods.
Main drawback
To recap, the main shortcoming of experimental approaches is that they cannot predict the effect of changes in cell culture media (which can have more than 50 components), feeding strategy, or process parameters on cell density or productivity. Considerable time and effort are spent in optimizing a process even as the potential for optimization is poorly understood.
Furthermore, cell culture process optimization has multiple objectives that are interdependent and compete with each other. For instance, there is often a tradeoff between productivity, cell density, and product quality. Together, these challenges make cell culture process optimization a complex problem that has a high-dimensional design space. Traditional approaches are unable to explore this design space thoroughly and therefore lead to suboptimal, non-robust cell culture processes.
Enter Digital Twins. Digital Twins of cell culture processes are capable of resolving the aforementioned challenges. A Digital Twin of a cell culture bioprocess mirrors a process of a real bioreactor. It can be used for predicting cell growth and metabolism under dynamic process conditions. Therefore, it can be used to run virtual experiments to provide critical decision support during process development.
How exactly can Digital Twins tackle the challenges mentioned above? First, Digital Twins can explore the high-dimensional design space of cell culture processes by virtual experimentation. They can predict the optimum media composition, feeding strategy, and other process parameters to achieve multiple objectives such as high cell density and productivity.
Second, Digital Twins explore the design space rapidly and systematically. For example, Digital Twins can simulate a 14-day bioreactor run in less than a second.
Third, a limited and predefined number of experiments suffice to build a Digital Twin that can better determine the potential for optimization. Finally, Digital Twins can evaluate the impact of process variability on critical quality parameters for process characterization.
Thus, the Digital Twin can develop an optimal and robust cell culture process to make biologics safer and more affordable.
Digital Twins
The integration of artificial intelligence with genome-scale biochemical models and bioprocess models has led to development of Insilico’s Digital Twins that offer predictive power and wide applications in bioprocessing.
Insilico’s Digital Twins consist of three basic building blocks (Figure 1). First, a model of a fed-batch or perfusion process that accounts for the dynamics in extracellular component concentrations due to feeding and sampling. Second, a model of the genome-scale biochemical pathways that provide the mechanistic foundation for simulating cellular metabolism via estimation of intracellular metabolic fluxes. Third, an artificial neural network (ANN) that learns the kinetics between the extracellular concentrations and intracellular metabolic fluxes. The ANN also learns the kinetics between extracellular component concentrations and process parameters that cannot be described mechanistically (for example, pH, temperature, vitamin concentrations, and trace metals).

Digital Twins integrate these basic building blocks to yield a high-quality predictive model of the cell culture process. In addition, Digital Twins have applications throughout the value chain in drug substance development.
At the development stage, Digital Twins can perform virtual experiments to advance cell line engineering, clone selection, cell culture process development, and process characterization. At the manufacturing stage, Digital Twins can be used for model-predictive control of the cell culture process.
Creating Digital Twins
First, routine time-course bioprocess data is applied to estimate the rates of cellular growth and substrate consumption. Then, the genome-scale biochemical model is utilized to estimate the steady-state intracellular fluxes at different phases of the cell growth. Next, the intracellular metabolic fluxes are resolved into elementary flux modes.
The elemetary flux modes represent the activities of linearly independent metabolic pathways that together can describe all the possible modes of cellular metabolism. The elementary flux modes can readily be transformed back to intracellular fluxes when required.
The next step is the bridge between the mechanistic models and artificial intelligence. Here, recurrent neural networks (RNNs), a class of artificial neural networks, learn the kinetics between the elementary flux modes and the extracellular component concentrations. The RNNs also learn the kinetics between the extracellular component concentrations and parameters that cannot be described mechanistically (for example, pH, temperature, vitamins, and trace elements). This step completes the development of the Insilico’s Digital Twins. It also makes a leap from steady-state analysis to kinetic analysis of the cell culture bioprocess.
Workflow: Creating process-specific Digital Twins
The Digital Twin needs to learn from experimental data to enable process-specific predictions. The training requires routine time-course data from bioprocesses such as cell counts, viability, and concentrations of amino acids, lactate, ammonia, and other components in the spent media. The training consists of two steps.
In the first step, historical data from the customer is used to preliminarily train the Digital Twin. The resulting preliminary Digital Twin is then used to create a model-based design of experiments (24 or 48 bioreactor runs, that is, one Ambr® run) with an objective to maximize the information content in the experimental data as this information-rich data is the basis for high predictive quality. In the second step, the data from the model-based design of experiment, carried out at the customer’s site, is used to train the Digital Twin, which is now ready for virtual experimentation.
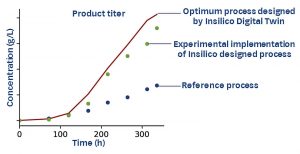
Here is an example of how Digital Twins optimize the monoclonal antibody titer of a fed-batch CHO-K1 culture process. In this case, while wet-lab experiments to increase the monoclonal antibody titer further were unsuccessful, the Digital Twin predicted an increase of 140% by virtual experimentation that tested thousands of feed media compositions and feeding schemes (time and volume of feeds). Experimental implementation of the optimized process achieved a 120% increase in monoclonal antibody titer from the reference process (Figure 2).
The Digital Twins can thus help complete process optimization in one round of bioreactor experiments. Furthermore, they can be transferred to customers for use in-house on additional molecules, and can be hosted on their servers or cloud services.
Future outlook: Factory of the future
Digital Twins bring together the best of mechanistic and data-driven modeling to enable automated predictions. While this article highlighted their application in the development of cell culture processes, they could play critical roles in the factory of the future, where they will be able to constantly communicate with bioreactors to gather data, predict performance, and if necessary, take corrective actions automatically and in real time.
This vision requires co-development of the Digital Twins and process control strategies, an endeavor that is currently under way.
Shilpa Nargund, PhD, is a Data Scientist & Business Development Manager, Kathrin Guenther, PhD, is a Sr. Business Development Manager, and Klaus Mauch ([email protected]) is the CEO & CTO at Insilico Biotechnology.


