Sponsored content brought to you by
Who has time to wait for a serendipitous discovery to occur? Good science starts with a good question. An unbiased approach to scientific research, like next-generation sequencing (NGS), can answer the questions posed as well as free experimental design from the bounds of prior understanding and preconceived expectations and uncover new findings.
NGS provides the power to sequence vast amounts of genetic material at a fraction of the time and cost of traditional methods.1 As the cost continues to decrease, this technology has become a critical asset to scientists. Now is the right time to leverage this impartial approach to reveal a broader landscape of molecular entities and enable deeper understanding of biological phenomena, pathways, and systems.
Easily Accessible Experts
An impactful effect of the genomics era has been the establishment and growth of core labs and service providers. Unlike in decades past, a researcher starting a new lab no longer needs to own all of the equipment that is generally needed for a given method. Core labs have all the instruments and knowledge in place to help build expertise and knowledge and often offer training and one-on-one consultations. Many also excel at facilitating advantageous connections between the wide swaths of scientists they service.
Regardless of your level of experience—from novice to expert—a core lab can help with either a portion or the entire process from experimental design to library prep, sequencing, and data analysis.
“The most beneficial thing about a core lab is that it is our job to be on the forefront of our area of expertise, and it is a scientist’s job to be on the forefront of their expertise. We can work together to advance science,” said Ann Tate, Project Manager, Transcriptional Regulation & Expression (TRex) Facility, Cornell Institute of Biotechnology. “Knowing what your question is as a scientist and just coming to us with that question is often a great first step because we can help you decide what kind of technology will work best for you and how best to use it.”
Charlie Johnson, PhD, Director, Genomics & Bioinformatics Services at Texas A&M University, summed it up by saying, “We like to do cool stuff with cool people.”
Core facilities perform a broad range of activities; some are standardized, but many are customized. They constantly tailor their procedures to the data the researcher wants to receive from the experiment.
For example, working in a core facility gives Adrian McNairn, PhD, Senior Research Associate, Genomics Innovation Hub, Cornell Institute of Biotechnology, an opportunity to both develop methods as well as help teach others how to use them.
“People can be ambitious when it comes to genomics because with the speed of the field, you can dream big and actually be in a position to obtain that goal, especially in collaboration with core facilities to have access to the equipment you would need,” McNairn said. “We want to be a space where people can come in, collaborate with us, get the training, have access to reagents, and actually get their projects off the ground and to the point where they have publishable data.”
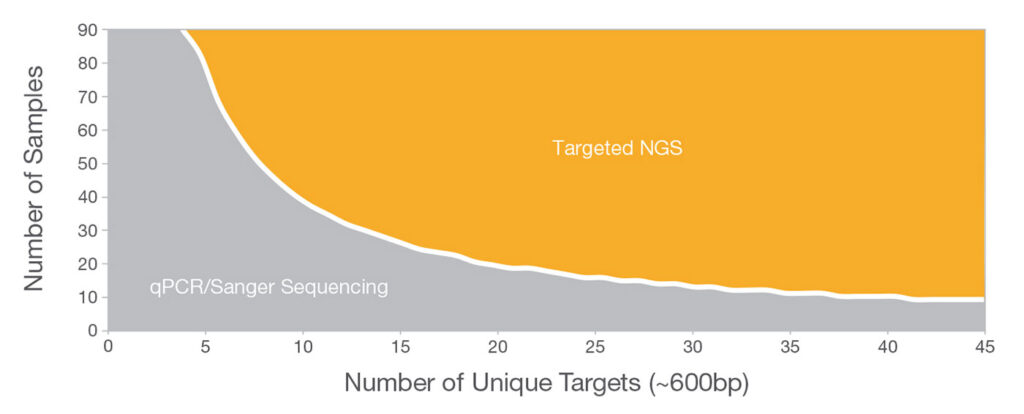
Thousands of Targets—Unique Discoveries
Initially, NGS often intimidates many scientists, from graduate students to postdoctoral fellows to principal investigators. Experimental design, sample collection, and analysis and interpretion of the vast amount of data that NGS provides appear insurmountable.
Both Mercedes Carro, PhD, a postdoctoral fellow, and Amanda Touey, a graduate student, study spermatogenesis in the Cohen Lab at Cornell University. Transcriptional regulation and dynamics are critical to understanding spermatogenesis. Part of the puzzle is the differential expression of Argonaute proteins that bind to small noncoding RNAs to regulate gene expression.
Neither came from a sequencing background nor had used NGS. In fact, Touey never thought that she would be studying RNA science and how it relates to reproductive biology.
“Talking to the Cornell Core Lab/Facility about our NGS goals and having them explain each step of the process made it more approachable,” Carro said. “Now I can understand why it is important to process your samples in a certain way and to have specific controls. You do not need to be an expert. For us, sequencing changes everything. Bulk RNA sequencing allowed us to see what was going on at the chromosomal level in our mutants.”
They also wanted to do small RNA sequencing. “The small RNA sequencing was going to take a lot of customization and things that were well out of the reach of our capabilities,” Touey emphasized. “The core lab became essential. They helped us through the protocol and how to get the best quality samples and were absolutely instrumental in helping us wade through all of the data from this megaproject so we could get something meaningful out of it. Something that seemed overwhelming when I first started grad school now seems attainable.”
For these researchers, NGS paved the way to acquiring unique and important data. With the assistance of the core facility, they were able to cast a wide net and look at data that otherwise would have been disregarded or thrown away.
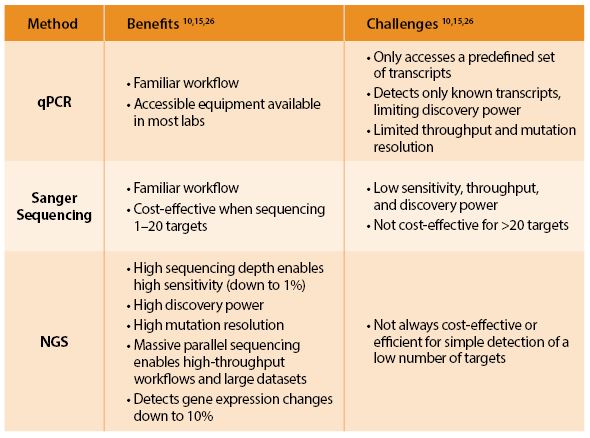
Novices Become Technology Embracers
Another postdoctoral fellow in the Grimson Lab at Cornell University, Louis Vu, PhD, was also inexperienced in NGS techniques, but wanted to use single-cell RNA-seq to take a broader look at the transcriptome of the immune system to get some insight into the mystery of myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS).
Vu feels that the Cornell Core Lab/Facility can help him and his colleagues move on their hypotheses more quickly by looking at questions in an untargeted way. “Whenever I come up with an NGS experiment, I discuss it with the Cornell Core Lab/Facility,” Vu said. “They might have previously analyzed samples similar to ours, and they can help with multiple steps of the workflow. They are also helping me understand more on data analysis and manipulation.”
Connor Kean, a graduate student in the same lab, is interested in epigenetic effects on the immune system. Since he was concerned that sample collection and processing would affect his results, he started working with the Cornell Core Lab/Facility early to guide experimental design. The Cornell Core Lab/Facility gave him confidence that once he prepared his libraries, he did not have to worry about quality control issues.
“I know a lot of labs that do almost exclusively biochemical assays, functional assays, and cell culture work, but are not necessarily thinking about ways in which genomic assays could be applied to their work and how they could facilitate the generation of some really interesting results,” Kean said. “If I could not access NGS, it would make my project extremely difficult because what I think is happening is that multiple loci are acting in concert to produce the phenomenon that we are seeing.”
Many pathways remain unexplained in multiple model systems—mammalian, microbial, plantal, avian, or insectile—and NGS can help open the door to a wider understanding. “Now is the time to get started, because the cost is right and because we have so much experience under our belt that we can facilitate any genomic experiment you want to do or can think about,” Tate said.
Reference
1. Wolff HB, Steeghs EMP, Mfumbilwa ZA, et al. Cost-Effectiveness of Parallel Versus Sequential Testing of Genetic Aberrations for Stage IV Non-Small-Cell Lung Cancer in the Netherlands. JCO Precis Oncol. 2022; 6: e2200201. doi: 10.1200/PO.22.00201.

To learn more on how to get started with NGS, download our NEW ebook at ilmn.ly/new-to-ngs-ebook-7.




