Sponsored content brought to you by
Alzheimer’s, Parkinson’s, dementia, amyotrophic lateral sclerosis (ALS) / motor neurone, and Huntington’s disease are examples of neurodegenerative diseases that have become increasingly important with high-profile campaigns raising awareness and focusing scientific effort. This research is gaining traction, with the understanding of these diseases evolving rapidly as exemplified in the Science paper by Balusu et al,1 which elucidates the mechanism of cell death in Alzheimer’s disease. However, the underlying cause of this disease still remains elusive. This research has, for Alzheimer’s disease, identified that fibril formation is the precursor to cell death via MEG3 (maternally expressed gene 3) upregulation. Other fibril types are also currently associated with not only Alzheimer’s disease, but also other neurodegenerative diseases, so identification of the specific fibril class and location within the brain is key to early and accurate diagnosis.
Understanding disease progression is a key area of research, but it requires identification of individual fibril types and their development over time. The preferred method is to use noninvasive, small-molecule tracers, or ‘biomarker’, compounds, which are selective for individual fibril types and enable early, accurate diagnosis and monitoring of disease progression to aid understanding and treatment development. The problem with fibrils is that they are not typical proteins. Typical proteins have a globular structure which can be processed and degraded naturally; fibrils are abnormal and once formed are difficult or impossible to degrade. They build up and lead to cell death, and this cell death is this cause of the symptoms associated with neurodegenerative diseases.
Fibrils are constructed from multiple individual protein fragments which align and pack into shelves which stack on top of each other. Unlike normal proteins, the overall structure of the fibril seems to be controlled by the local environment, which makes absolute fibril structural determination difficult. Structural determination of the TDP-43 fibril has yielded several distinct forms. Even those extracted from Alzheimer’s patients show differing structural forms. There are currently marker compounds available and new compounds under development, but there are problems and difficulties associated with their development such as brain penetration, fibril selectivity, and toxicity, so the design of new classes of compounds is required.
Addressing this design problem computationally is challenging with structure-based drug design tools, as these methods require both a defined protein structure and a known binding site about which to inform an investigation. For fibrils, there are multiple defined protein structures, with very little information on ligand binding.
Cresset Discovery has developed a ligand-centric approach which removes the requirement for knowledge of the ligand-binding mode and even the protein. The method instead uses known ligands to identify key requirements for binding with or without knowledge of the protein. This information is then employed to identify additional ligands which also match these requirements. However, unlike exact matching searches, the Cresset field-based technology is a more abstract method that approximates ligands to a set of fields and field points which represent the ligand’s electronic, lipophilic, and steric environment and is therefore chemically independent. This chemical independence allows for the identification of new compounds that are biologically similar but chemically differentiated from their parents, which means that development problems are not inherited by these newly identified compounds.
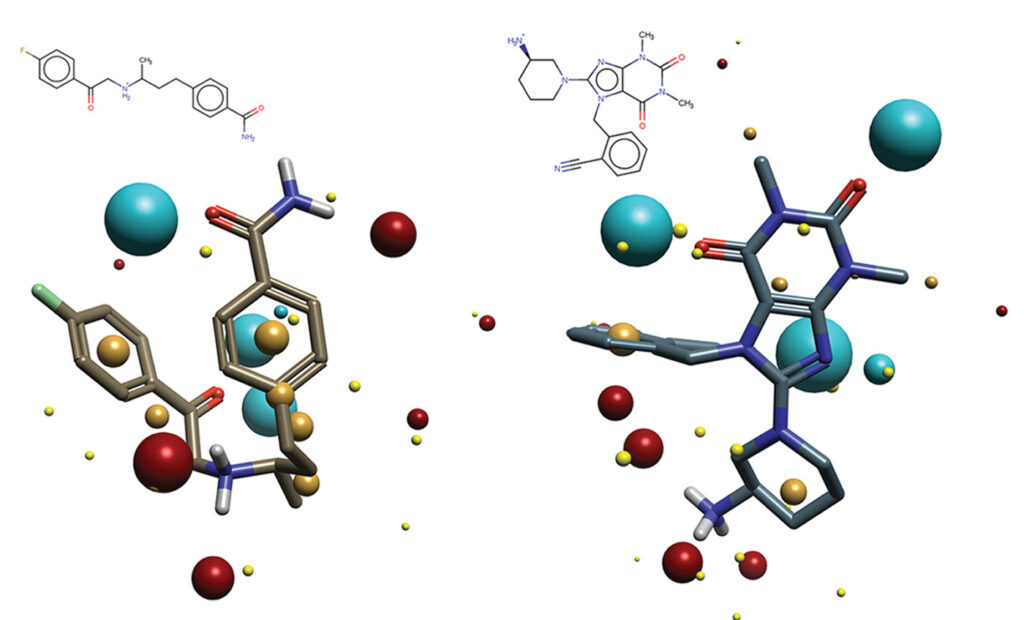
Figure 1 shows an example of how using Cresset field-based technology can identify very diverse chemistries which share biological actives.
Cresset Discovery has applied this technology successfully across multiple target classes including those relevant to neurodegenerative diseases. Learn how Cresset Discovery offers a complete solution that combines specialist software tools with molecular design expertise that can advance your project development.
References
- Balusu et al. MEG3 activates necroptosis in human neuron xenografts modelling Alzheimer’s disease. Science 2023; 381: 1176–1182.
- Dickmanns A, Damerow S, Neumann P, et al. Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major. J. Mol. Biol. 2011; 405(2): 461–478. DOI: 10.1016/j.jmb.2010.10.057.
 Learn more about Cresset Discovery’s intelligent in silico discovery approaches www.cresset-group.com/discovery
Learn more about Cresset Discovery’s intelligent in silico discovery approaches www.cresset-group.com/discovery



