In Vitro–In Vivo Extrapolation (IVIVE) is a method that allows researchers to predict the rate at which substances are metabolized within the human liver and are then eliminated from the body. IVIVE is compelling because it offers the ability to use in vitro tests to determine pharmacokinetic impact in vivo.
In vitro research provides high throughput, rapid results, and cost efficiency, which is why it remains the primary methodology in new compound discovery. But in vitro testing cannot fully reflect a compound’s characteristics the way in vivo methods can. And understanding the structural transformation a compound undergoes inside the body is critical when estimating dosage and optimizing compounds in clinical research.
By providing a pathway to predict in vivo data through in vitro means, IVIVE has the potential to deliver the best of both worlds. Predicting clinical utility from preclinical data is not just scientifically relevant, it is also business savvy. Developers can use IVIVE to understand and quantify the relationship between external and internal exposure and toxicological response to substances. This data can facilitate future drug discovery, reduce the time and cost of developing new products, and inform chemical risk assessments. Put simply, IVIVE can streamline drug development—as long as developers use the right model.
Building an IVIVE model
Establishing an IVIVE model requires two parts: in vitro liver clearance data and a correction equation used to extrapolate data in vivo. The first part uses a relatively simple metabolic model to determine the rate at which human liver cells (that is, hepatocytes) and liver subcellular fractions (that is, microsomes) metabolize substances.
The key to achieving accurate clearance data is optimizing in vitro metabolic assay protocols. High metabolism rates can present significant challenges for new discovery compounds. But a robust, stable in vitro metabolic assay can help rank order compounds and improve new compound design. It can also provide a method for predicting in vitro data in vivo. Building an effective in vitro model is critical to success with IVIVE, so developers who don’t have internal resources or expertise should consider working with a laboratory partner.
The second part of building an IVIVE model—establishing the correction equation—is more challenging, but it is the primary goal of the research. Establishing the correction equation helps accurately predict human clearance and requires comparing three types of data—measured, accepted, and theoretical. As with any predicted data, there is potential for error.
A step-by-step guide to improving in vivo prediction
Despite wide usage and documented success, IVIVE technology is prone to underestimation. In vitro prediction in vivo is underestimated with a 3- to 10-fold systematic error. Researchers have worked tirelessly to optimize the existing models and reduce systematic error, but underestimation still exists, and the mechanism is unclear.
So, the question remains: How to further optimize the metabolic reaction system in vitro to establish the corrected in vitro intrinsic clearance? Few laboratories have developed assays and data analyses that are advanced enough to accurately predict human clearance. But the steps below can put developers on the right path to doing so.
Step 1. Optimizing the metabolic reaction system
Research using an optimization assay has shown that it is possible to significantly improve clearance in vitro and reduce underprediction in vivo. But IVIVE does not end there.
Compounds will bind to liver microsomes and hepatocytes when incubated in vitro. Theoretically, only unbound (that is, free) compounds can be metabolized. So, developers need to account for binding when calculating liver intrinsic clearance.
If done correctly, optimizing the in vitro system will increase the rate of metabolism and reduce the underprediction of in vivo clearance.
Optimizing the in vitro assay requires the following actions:
• Increase the shaking speed in hepatocyte incubations to minimize any unstirred water in the assay.
• Increase the ratio of organic solvent to reduce the apparent stability of insoluble compounds.
• Select a suitable incubation time and protein concentration.
Once the in vitro assay is complete and researchers capture liver intrinsic clearance data, the IVIVE process can begin. It starts with using commercial compounds whose human pharmacokinetics are already known. Compounds are chosen based on three factors: 1) adherence to the liver metabolism model; 2) representative range (that is, whether the compounds are acidic, alkaline, or neutral); 3) documented clearance data.
The compounds are subjected to in vitro human liver microsome and hepatocyte stability experiments to establish a measured intrinsic liver clearance rate. Calculating the difference between in vivo and measured in vitro intrinsic liver clearance rates helps developers establish the correction equation in Step 2.
Finally, developers can use binding assays to measure the interaction between the chosen compounds and the liver microsomes and hepatocytes. A third assay focuses on protein binding in human plasma. Plasma protein binding needs to be accounted for. The final data point collected should be the drug distribution ratio of human whole blood to plasma.
Step 2. Establishing the correction factor
The equation used to establish the relationship between in vitro and in vivo clearance is called the “well-stirred model.”

It is one of the simplest and most widely used predictive models, especially when conducting early screening for new chemical entities.
The well-stirred model assumes that the diffusion rate of drug into liver is not limited by any barriers. Only unbound drugs cross the cell membrane and occupy enzyme sites. Metabolizing enzymes are considered to be evenly distributed in the liver. For any organ, the in vivo clearance rate is determined by three factors: blood flow of organs (Qh); the unbound fraction of compound in blood (fu); and the intrinsic clearance of organs to drugs (CLint).
The well-stirred model can help predict in vivo clearance.
And if the measured data of in vivo clearance is known, the model can also help calculate the theoretical liver intrinsic clearance in vitro (Equation 2).

Fifteen commercial compounds with measured in vivo human pharmacokinetic clearance were selected for in vitro human liver microsome and hepatocyte stability assays. Then the theortcical CLint(liver) was calculated and the calibration equation of liver intrinsic clearance was established by a linear regression model: Theoretical CLint(liver) = (a × measured CLint(liver)) + b.
This correction equation is the primary goal of IVIVE research—it can be used to correct in vitro liver intrinsic clearance rates in future testing. This method was used to achieve an underprediction of 1.25-fold for the hepatocyte assay and 3.5-fold for microsome stability assays.
Step 3. Applying the correction equation
The correction equation serves as the framework for accurately predicting in vivo clearance rates. The process looks like this:
• Run the in vitro metabolic stability assay to get the in vitro intrinsic clearance (CLint(liver))
• Correct the in vitro intrinsic clearance using the established correction equation (CLint(liver)-corrected)
• Use the corrected in vitro clearance (CLint(liver)-corrected) to predict the in vivo clearance using the equation below.
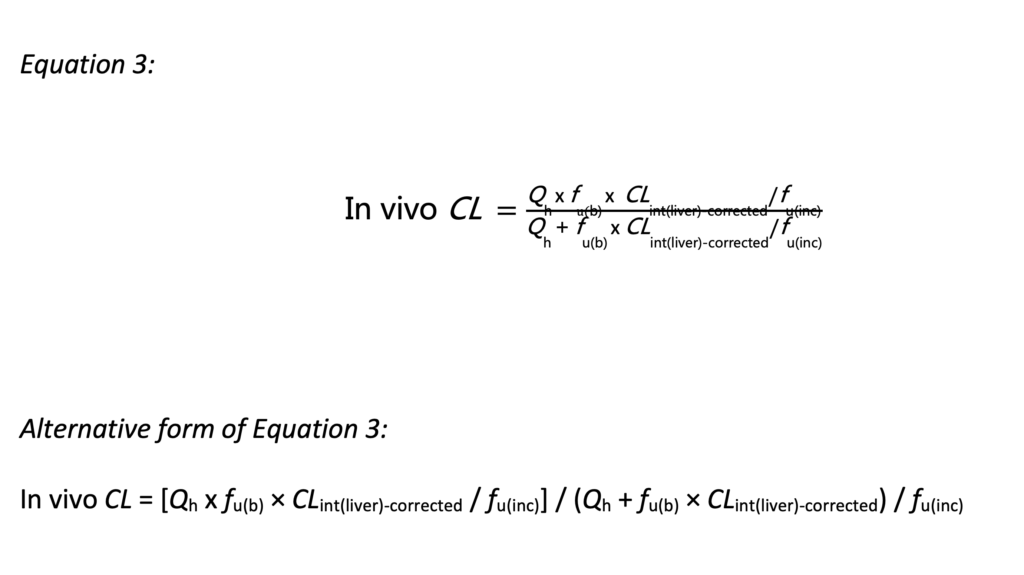
Using the equations in this article, it is possible to assess the underprediction of clearance rates in hepatocytes and liver microsomes.
The bottom line on IVIVE
IVIVE is a simple and fast way to predict in vivo clearance rates using in vitro data. For these reasons, IVIVE is especially suitable for compounds in the early discovery phase that demonstrate the potential for profound clinical applications.
Xiangling Wang, PhD, is a director and the head of the platform for drug metabolism and distribution at WuXi AppTec.


Comments are closed.