Mass spectrometry (MS) has become the single most significant method for identifying proteins, lipids, metabolites, oligonucleotides, and even supramolecular analytes. MS has also become increasingly quantitative. The hard numbers that are generated by MS, in combination with spectral databases, can yield precise answers for research institutions and biomanufacturing organizations alike.
In biomanufacturing, MS can accomplish the analysis of cell culture contents, the determination of critical process parameters, the implementation of quality control measures, and many other tasks. And MS continues to become more capable—as well as more accessible. Indeed, if we review several recent MS developments from the past year, we can see that the technology’s contributions to biomanufacturing are far from exhausted.
Simplified proteomics
For example, in November 2021, Sciex entered an agreement with Evosep to co-market products “to improve robust, high-throughput proteomics workflows for precision medicine.” Central to this effort are the Sciex ZenoTOF 7600 mass spectrometer and the Evosep One high-performance liquid chromatography (HPLC) system.
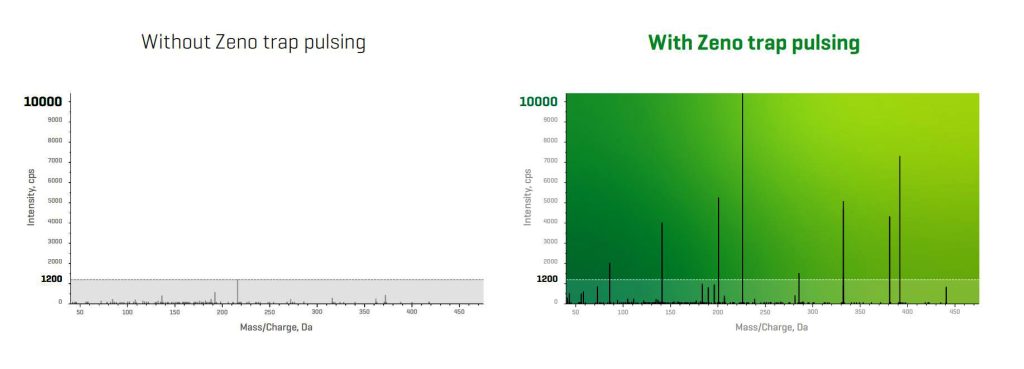
Evosep One employs single-use trap columns to desalt samples offline, an approach that reduces sample handling and injection cycle overhead. Disposability eliminates cross-contamination. ZenoTOF incorporates electron-activated dissociation (EAD), which exploits several electron-based fragmentation mechanisms within one device and extends analytic capabilities to all molecule types, from singly charged small molecules to multiply charged proteins.
According to Jose Castro-Perez, the senior director of the Accurate Mass Product Line at Sciex, proteomic workflows are a minefield of potential technical problems. “Some of the biggest challenges,” he says, “include depth of protein identification and quantification, utilization of instrument time to maximize usable MS/MS coverage, dynamic range for both labeled and label-free quantification, throughput sufficient for large cohort studies, and localization of labile post-translational modifications.”
Castro-Perez says that the Zeno trap increases the duty cycle to 90%, improving sensitivity at the MS/MS level for proteomics studies by factors of between 5 and 20. “The Zeno trap,” he adds, “allows more proteins to be quantified at much lower concentrations, for example, in labeled single-cell proteomics.”
When promoting the ZenoTOF/EAD system, Sciex underlines its ability to address modern medicine’s “developability gap.” Castro-Perez defines this term as the uncertainty arising from information deficits during development.
“Developability is about turning good candidate molecules into therapies,” he explains. “Many candidates show excellent potency, but not every molecule can be produced in the yields and with the physicochemical characteristics required to bring it to market.” Bridging the development gap therefore requires tools that provide understanding of both molecule and process, early in development, to support a “fail fast” strategy.
“Our customer feedback emphasizes higher-throughput proteomics and deeper proteome analysis,” Castro-Perez tells GEN. “[This is evident] not just in the number of proteins we can identify within a matrix, but also in the precision with which we can quantify these proteins.”
MAMs all the rage
Thermo Fisher Scientific was busy at last year’s American Society for Mass Spectrometry conference, introducing what it termed “new generation” MS instruments, software, and workflows. Among them were the Thermo Scientific Orbitrap Exploris MX mass detector, which enables multi-attribute methods (MAMs) for analyzing intact monoclonal antibodies, oligonucleotides, and peptides. The system also simplifies analytical technology transfer by allowing customers to use methods established during discovery or development to support manufacturing. According to Thermo Fisher, the system promises “high-confidence results, even for users with limited mass spectrometric experience.”
On the software side, the Thermo Scientific MAM 2.0 workflow streamlines molecule characterization, process monitoring, and release testing. It does so by replacing multiple traditional quality control assays with one comprehensive analysis throughout the molecule’s lifecycle. Workflow analyses are supported by Thermo Scientific Proteome Discoverer 3.0 software, which incorporates the CHIMERYS (by MSAID) peptide identification artificial intelligence (AI) engine.
“MAMs require high accuracy and the ability to detect more than one species at once,” says Andreas Huhmer, PhD, global marketing director for proteomic MS at Thermo Fisher Scientific. “Orbitrap MS makes this possible because of its ability to measure small mass differences. Standard MS must be calibrated within 24 hours, but not Orbitrap.”
Orbitrap also facilitates the development of lifecycle-proof methods that support both development work and quality control. “Extensive method development normally occurs during product development,” Huhmer notes, “but assays are usually redone to meet the needs of production-related activities.”
During initial method development, Orbitrap Exploris MX collects data in both MS1 and MS/MS modes. During discovery/development, MS1 provides precise masses from peptide mapping, whereas the MS/MS spectrum gives the sequence.
“MS/MS detects subtle changes to the backbone, including post-translational modifications,” Huhmer details. “This information is not routinely required during production, but the MS1 continues to be used to determine precise masses. You no longer have to worry about these changes because they have already been quantified and characterized, and the process is set. Since the objective now is product purity and process stability, detailed MS/MS assays are not needed.”
Software plays a critical role as well in transferring a method directly from development to quality control. “All MS data goes to the same repository, so you can access the data and method anywhere on earth,” Huhmer explains. He sums up method transfer as follows: During development, assays are created to generate critical quality attributes that are “consumed” in quality control.
CHIMERYS is not essential for MAM workflows. Rather, it is advantageous in research and discovery, where it can help resolve spectral overlaps.
“There’s so much spectral overlap from some samples,” Huhmer says. “Up to 15 different peptides may co-elute as a single peak, which cannot be resolved through database searches. AI knows not only masses, but relative intensities of fragmented ions. It also knows what the MS/MS fingerprint looks like for a specific sequence, which often is not the one predicted theoretically. AI anticipates those differences, even for spectra that are not in a database.”
Improved decision-making
In February 2022, Agilent Technologies acquired AI software, patents, personnel, and related assets from Virtual Control, an AI and machine learning software developer. Agilent will incorporate the software, ACIES, into its MassHunter applications for both LC-MS and GC-MS systems to improve the productivity, efficiency, and accuracy of these systems in high-throughput laboratories.
ACIES automates labor-intensive GC-MS and LC-MS data analysis tasks by streamlining the analysis workflow from sampling to reporting.
We normally think of AI as a decision-making or algorithm-generating tool, so the connection with sample preparation is not obvious. Thomas Bispham, associate vice president, Analytical AI/Machine Learning, Agilent, explains that ACIES falls more precisely within machine learning, a subdiscipline of AI. “This AI/machine learning technology will improve efficiency in high-throughput lab workflows,” he asserts. “Incorporating digitalization into GC-MS instruments positively impacts the complete workflow and, ultimately, the entire value delivery channel.”
Solving complex complexes
In 2018, Indiana University professors Martin Jarrold, PhD, and David Clemmer, PhD, founded Megadalton Solutions to commercialize charge-detection mass spectrometry (CDMS). In early 2022, Megadalton’s CDMS-relevant assets were acquired by Waters. Now the technology will be developed within Waters’ MS portfolio. CDMS facilitates analysis of very large proteins and protein complexes, such as those used in gene therapies, which are difficult to analyze with conventional MS due to their chemical complexity.
According to Steve Preece, PhD, principal consulting product marketing manager at Waters, large molecules and complexes can be quite heterogeneous and “adducted,” resulting in broad mass distributions. “When adducts are convoluted with the large number of distinct charge states during ionization,” he says, “broad envelopes of unresolved charge entities occur that span a range of mass/charge (m/z) values.
“Under such conditions, data acquired with typical MS systems, which measure m/z values, cannot be used to determine the analyte mass as charge states cannot be assigned. The enabling feature of CDMS is that it simultaneously measures both the m/z and z of individual ions, allowing direct mass determinations.”
CDMS has been around since the 1960s, primarily as an academic technique. While it has shown potential, only recently has it proved capable of solving challenges in the broader market. “Recent approvals of gene-based therapies have stimulated the demand for a commercial product to support development of these therapies,” Preece adds.
CDMS works on a broad range of extremely large “molecules” such as gene therapy vectors, viruses, vaccines, lipid nanoparticles, and exosomes. “The highest-profile application right now is the measurement of adeno-associated viruses (AAVs), small viruses that are used to deliver single-stranded DNA to cells,” Preece says. “Since we need to know whether the AAV capsid contains the required payload, it is necessary to measure the relative amounts of empty, partially full, and full capsids as critical quality attributes. The technique also has the potential for analyzing very large protein complexes, in the megadalton range, that are outside of the capabilities of traditional MS.”
Beyond biomolecules
Increasingly, MS supports molecular characterization outside of direct biomolecule quantitation. For example, MS has been used to analyze contaminants that do not directly arise through cell culture.
In February 2022, MilliporeSigma and Waters announced that they were collaborating on an MS reference library for extractables and leachables (E&Ls), which are contaminants of concern in single-use bioprocessing. The companies say the library will enable analytical labs to identify potential E&Ls in their samples by using Waters’ ion mobility–enabled LC-MS instruments and then confirming the identity and quantity using Supelco reference materials. The companies noted that they were interested in moving the industry closer to consensus on reportable E&L thresholds, control options, and pharmacopoeial standards.
Bioprocessors have been concerned about E&Ls since the early days of single-use manufacturing, yet they have yet to agree on which parameters are most relevant or how they should be assessed.
“Harmonization is difficult since there are several thousand E&L compounds and diverse regulations in different countries and regions,” notes Markus Obkircher, PhD, head of reference materials and proficiency testing R&D at Merck Life Science. “Trace analysis of these materials is also challenging due to potential cross-contamination from sample preparation or from instruments and tubing. USP <1663> and PQRI recommendations provide excellent guidance for LC-MS or GC-MS E&L screening methods.”
The Supelco standards provide reference materials for rapid presence/absence screening, as well as for accurate quantification of E&Ls. Customers can choose either neat materials or ready-to-use mixes. Each has its advantages.
For the development of neat compounds, quantitative NMR (qNMR) is the certification method of choice since it offers direct traceability to the SI unit through primary standards. Alternately, mixtures offer greater ease of use. A novel approach involves combining qNMR with isotope dilution MS to offer reference materials in solution while still assuring homogeneity and stability throughout the product’s shelf life.
Detecting E&Ls is normally achieved through LC-MS, but the challenge is screening for many structurally diverse compounds, arising from various matrices, in one chromatographic run. “A further difficulty,” Obkircher points out, “is the presence of known-unknowns or unknown-unknowns in a sample.”
The method parameters for LC-MS systems are complex, and the detection of multiple analytes is more challenging than for GC-MS. Different source designs, ionization methods, matrix effects, and eluent systems need to be considered to perform a suitable analysis. “In that context,” Obkircher advises, “the use of certified reference materials is an ideal way to check for such matrix effects by dilution within the matrix.”
Packaging and barrier materials serve as a first-line defense for drug quality, but the increasing capabilities of analytical science have uncovered some inconvenient truths, namely, that the materials themselves represent a contamination risk. Hence the interest in E&Ls.
Conclusion
Technologic improvements have led to the democratization of MS and its widespread adoption in many industries. For the life sciences, this transformation has led to an explosion in methods. For the biopharmaceutical industry, MS has become a go-to method employed from early discovery to post-marketing surveillance.
“Mass measurements are the most direct evidence for what is in the product,” says Thermo Fisher’s Andreas Huhmer. “Ultimately, if you want to understand biology, you must be able to quantify sometimes subtle differences among molecules, which is difficult by any method other than MS.”