January 1, 2015 (Vol. 35, No. 1)
Exploring Cancer’s Signature in the Sensitive Functional Domain of the Human Proteome
Lung cancer is one of the most common malignancies and the leading cause of cancer-related fatality. In the U.S. alone, more than 210,000 new lung cancer cases are diagnosed every year, with more than 170,000 deaths resulting from the disease each year.
Current diagnostic practices for common cancers rely heavily on imaging technologies such as CT scans for lung cancer, mammograms for breast cancer, and pelvic ultrasounds for ovarian cancer. Given the high probability of false-positive findings associated with CT screening, there is a substantial need for additional noninvasive modalities to discriminate between benign and malignant nodules. There are similar challenges in imaging-based screening for other malignancies and a subsequent need for complementary diagnostic tests.
Blood-based biomarkers have potential in cancer screening, and their role could extend further from general population risk assessment to treatment response evaluation and recurrence monitoring. Despite a large literature collection related to biomarkers for common cancers, blood-based diagnostic tests that inform about the presence of cancer at an early stage and predict treatment response have been difficult to develop.
Protein markers currently in clinical use have limitations with respect to their use for screening owing to low sensitivity and specificity in early stages and inability to distinguish aggressive from indolent tumors. Other common cancers, notably breast and lung cancer, lack established biomarkers with demonstrated clinical utility in a screening setting. Thus, there is a need for biomarkers with the required sensitivity and specificity for the detection of frequently occurring cancer types.
Functional Annotation
Most efforts in proteomics seek to identify and sequence annotate the proteome by LC-MS/MS analyses of peptides derived through proteolytic processing of the parent proteome. Thousands of proteins have been identified from human serum as a result. However, little attention has been paid to functional annotation. Yet functional annotation is crucial as the landscape of protein conformations is highly variable, each conformation may contribute to its own functional activity.
Sequence annotation alone cannot capture this vital information, so new strategies are necessary. The challenge has been to overcome the analytical bias toward the most abundant proteins, and the complexities of mining the data to a manageable number of biomarker proteins that can be analyzed in more depth.
So reconciling protein identifications to actual enzyme activities or functions has been subject to limitations in proteome separation and assay technologies. To overcome these inefficiencies in functional annotation, a top-down approach, starting with function, and ending with sequence and structural annotation is now available. The PEP technology, developed by Array Bridge, uses modified 2D gel electrophoresis to separate the proteome, without substantially compromising function. The isolated proteins are then electro-eluted from the PEP plate, and enzyme activities are measured systematically.
This method thus provides a new functional dimension to explore the human serum proteome. It is hypothesized that the levels and distributions of certain enzymes could reflect physiological changes of an individual and serve as possible biomarkers or diagnostic parameters. Our initial studies using lung and breast cancer patient serum and normal serum provided strong evidence for a clear disease signature when the enzyme activities are compared.
It is anticipated that this novel technology will provide a fundamentally new approach for breast and lung cancer diagnosis. The technology can easily be extended for the development of biomarkers, drug targets, or diagnosis kits for other types of cancer or disease.
Because of the limited loading capacity in 2D gel electrophoresis, high-abundance depletion with consequent low-abundance protein enrichment is desirable.
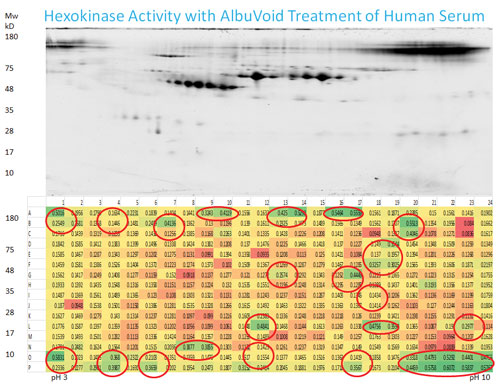
Figure 1. Human serum was first enriched by AlbuVoid for nonalbumin proteins, after 2D gel separation and protein refolding; hexokinase activities were analyzed from the eluted fractions on the PEP plate.
Proteome Enrichment
Biotech Support Group has developed a series of suitable functional proteome enrichment products that have been adapted into the PEP workflow. In Figure 1, AlbuVoid™ was used to enrich nonalbumin proteins from the serum before the PEP functional analysis. Figure 1 shows that the number of hexokinases and their activities were increased significantly after AlbuVoid enrichment. Because of the powerful separation capability of 2D gels and the effective refolding and elution of the active proteins after gel electrophoresis, a large number of enzymes could be analyzed simultaneously and produce an enzyme activity map for easy analysis.
Figure 2 is an example of analyzing hexokinase activities from both normal and lung cancer patient serum. As can be seen, both the distribution and relative hexokinase activities are very different between the two groups. It has been shown from another study that the fractions with enzyme activity could be recovered and proteins of interest could be identified by mass spectrometry (paper submitted).
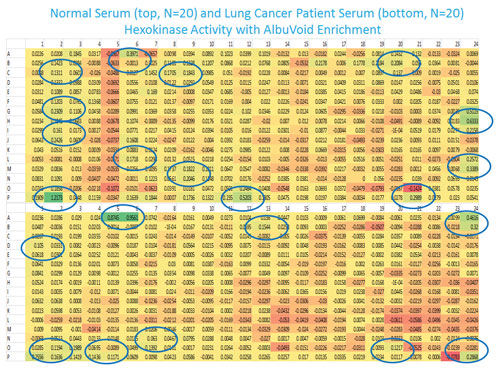
Figure 2. Human serum from 20 normal or lung cancer patients were mixed equally and analyzed for hexokinase activity after protein enrichment with AlbuVoid.
It is believed that a selected group of enzymes from the same family or different enzyme family could serve as biomarkers for cancer and other diseases after the further validation of the enzyme activity and development of specific monoclonal antibodies for the selected enzyme group, a flow chart on the process of function-based biomarker discovery is shown in Figure 3.
Using the PEP technology in combination with a consumable serum protein enrichment method like AlbuVoid, much more information could be extracted from the human serum.
The PEP technology is an open platform adaptable to any measurable protein function. It serves as an efficient method for high-sensitivity proteome characterization, quantitative and systematic top-down analysis of isolated protein function, and purity suitable for mass spectrometry protein identification.
In addition to the study of functional serum proteins, this technology can be used for pattern profiling of large enzyme families such as protein kinases and proteases, and any protein function with multiple variants from cells or biological tissue. In different tissues, or under stress or disease conditions, protein sequences that are normally associated with one function, may perform alternative functions—that is what we call the multifunctional proteome.
This multifunctional proteome can now be monitored, catalogued, and annotated by this PEP-based functional proteomic strategy. Using a top-down approach, we can start with any measurable protein function, resolve functionally active proteins into microwells, and from there, sequence and structurally annotate the subset proteome that presents that function. It is believed that the functional analysis of a proteome, in combination with other system biology approaches, will provide further insights into important biological processes and human diseases.
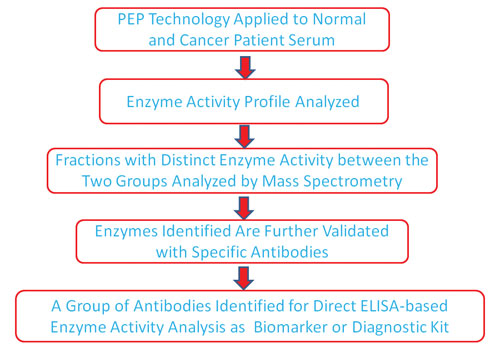
Figure 3. Flow chart of PEP technology for cancer biomarker discovery.
Zhenyu Sun, M.D., is chief surgeon at Nantong University School of Medicine Wuxi No. 3 Hospital. Xiaofeng Chen, M.D., is chief surgeon at Shanghai Huashan Hospital, Fudan University School of Medicine. Xiong Su, Ph.D., is a professor at Soochow University Medical College, China. Gan Wang, Ph.D., is an associate professor at the Institute of Environmental Health Sciences, Wayne State University. Matthew Kuruc is vice president for business development at Biotech Support Group. Xing Wang ([email protected]) is president of Array Bridge.



