
Product Manager
Bio-Rad Laboratories
The recently published recommendations of the European Commission Joint Research Centre (JRC) shine a spotlight on the versatility and scientific benefits of universal antibody libraries in combination with selection technologies, such as phage display, in generating precision antibodies for biomedical and scientific use.1 The JRC Science for Policy Report reveals a need for broader understanding across the scientific community of the disadvantages of animal-derived antibodies in terms of reproducibility, specificity, and flexibility, and challenges the widely held historical belief that animals are the only source for novel antibodies.
The report documents the findings of the European Union Reference Laboratory for Alternatives to Animal Testing (EURL ECVAM), which has a mandate, granted by EU legislation, to ensure the protection of animals used for scientific purposes. The EURL ECVAM explains that, aside from the ethical concerns associated with the use of animal-based techniques when alternative methodologies exist, the reliability of animal-derived antibodies is known to be often seriously flawed. In contrast, non-animal-derived molecular antibody tools offer greater precision, reproducibility, and flexibility.
The review states that animals should no longer be used for the development and production of antibodies for research, regulatory, diagnostic, and therapeutic applications. Instead, large antibody libraries and in vitro selection methods should be leveraged to address the shortcomings of conventional animal-based approaches.
The current situation
Despite the widely acknowledged drawbacks associated with traditional antibody generation techniques (for example, time-consuming protocols, quality issues, and poor reproducibility), polyclonal and monoclonal antibodies are still made by immunization of laboratory animals. Methods used to generate animal-based antibodies have not improved significantly over recent decades. Problems associated with lack of renewability and specificity, unknown genetic sequences, and the loss of hybridomas for monoclonal antibodies result in significant and costly wastage of materials and time.2 There is a general acceptance across the scientific field of the intrinsic limitations associated with animal-based antibodies. The JCR report creates a demand for scientifically superior resources.
Many non-animal-derived antibodies are already approved in Europe and the United States for use in therapeutics and diagnostics, and many thousands have been generated as research reagents. Indeed, academia and biotechnology companies have developed precision antibody products from non-animal sources for more than 25 years, some of which are widely commercially available and have been well characterized and validated in applications.
Leveraging existing expertise and established technologies
The JCR report recognizes the vast and robust body of evidence supporting the use of non-animal-derived affinity reagents as well as the practical advantages that these products offer concerning consistency and reproducibility. Laboratory-based techniques allow protocols for the generation of antibodies and antibody fragments to be standardized, refined, and optimized.
Universal antibody libraries were developed approximately 30 years ago and have been improved over time. Large libraries provide multiple benefits, including high success rates and rapid antibody generation. For example, the fully synthetic Human Combinatorial Antibody Libraries (HuCAL®) phage library, used by Bio-Rad Laboratories to generate recombinant antibodies for research and diagnostics and for the company’s catalog products and custom service offerings, contains 45 billion functional human antibody genes in Fab format.3
The JCR report recommends the use of guided in vitro techniques for selection of antibody sequences defined by essential characteristics (for example, stability and cross-reactivity). As the most mature technology in this field, phage display is already widely used and capable of efficiently generating a near-infinite range of high-quality antibodies from such libraries (Figure 1). Antibodies generated through phage display libraries may be developmentally, functionally, and structurally equivalent to those produced by animals, and improvements can be made to elevate quality further by refining or adapting the resulting gene sequences.4
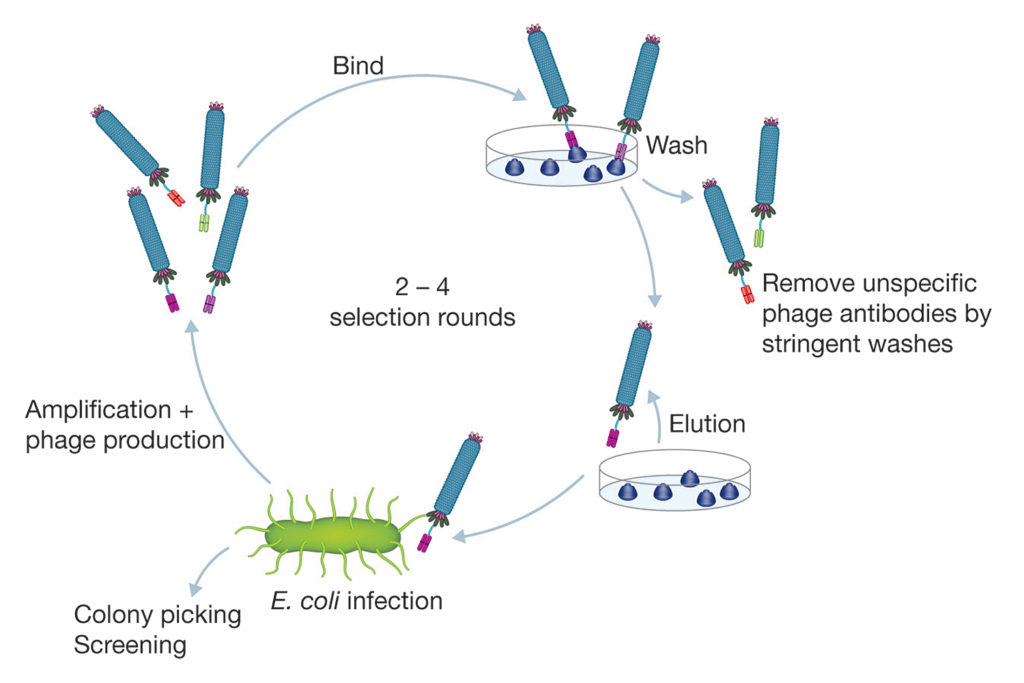
Universal libraries and in vitro selection strategies offer a powerful combination, enabling isolation of antibodies against virtually any type of antigen. These may include low immunogenic or “self” antigens and toxins, or antibodies with the ability to differentiate between two subtly different proteins. The resulting recombinant antibodies can be further engineered to offer the flexibility of different formats, such as Fab and bivalent Fab, constructs that may incorporate a variety of detection and purification tags, and full-length immunoglobulins of any isotype. They can also be further engineered to gain incremental improvements in terms of binding affinity by molecular evolution.
The JCR report states that a universal recombinant library can produce a diversity of antibody candidates equivalent to a lifetime supply of animals. Indeed, it is easy to see how the use of bench-based methods can offer considerable economic, resource, and time efficiencies when compared with the cost of ongoing animal care, immunization, and antibody purification procedures—in addition to the frequent losses due to low product yield or low quality associated with polyclonal sera or hybridoma cells. Protocols that use synthetic libraries and phage display selection platforms allow rapid generation and screening of novel antibodies in a matter of weeks, whereas animal-based methods take many months to complete.
A look to the future
If stakeholders in the scientific community step away from the widespread use of animals to generate affinity reagents, the resulting increase in demand for non-animal-derived products will drive further technological enhancements and innovations in this arena.
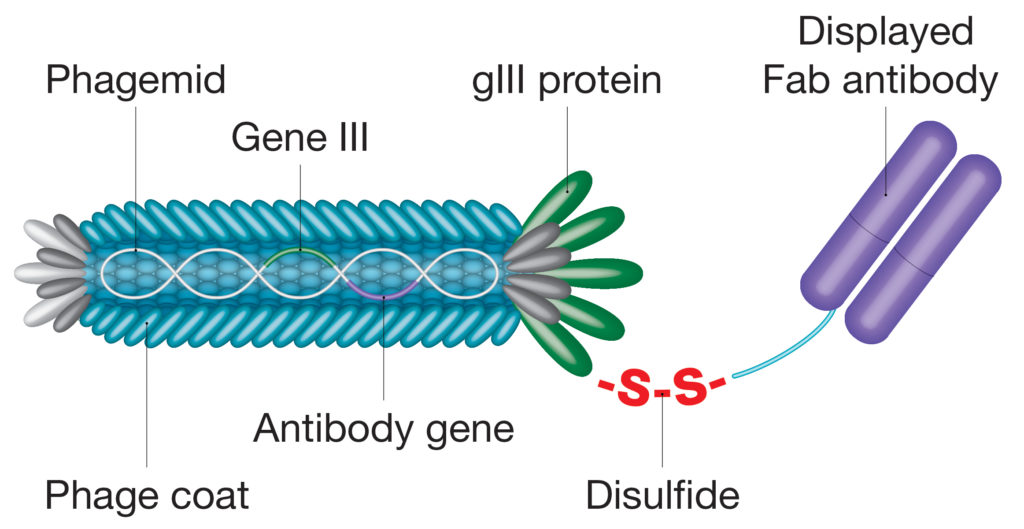
Evolving proprietary phage display systems are already bringing new possibilities and enabling antibody selection processes to be simplified and streamlined. For example, the CysDisplay® proprietary platform uses an engineered disulfide bridge to link antibody fragments to a phage protein, allowing elution through the addition of a reducing agent, rather than relying on traditional affinity-dependent approaches (Figure 2).5 This feature allows for better automation and retrieval of very high-affinity clones. Advances in protein ligation technologies have seen sophisticated molecular tagging tools being incorporated into recombinant antibodies, such as the SpyTag/SpyCatcher system, that circumvent cloning steps or facilitate antibody site-directed conjugation.6
Moving forward, existing, well-characterized monoclonal antibodies from hybridomas should be sequenced and produced using recombinant systems. To tackle the issues associated with the production of polyclonal antibodies, the EURL ECVAM panel recommends laboratory-based methods that generate defined combinations of recombinant antibodies (or “multiclonal” antibodies) from universal phage display libraries. This has the potential to combine the advantages of polyclonal antibodies (in terms of multiepitope binding and improved sensitivity) with the consistency of recombinant antibodies, and to provide a renewable resource.
A call to action
The JCR report concludes with a call for greater education, training, and awareness concerning the potential of non-animal-derived antibodies. Activities should focus on easing access to reagents and communication of case studies that emphasize the quality of these products. This needs to be driven by government authorities, funding agencies, and publishers through the setting of higher scientific, regulatory, and ethical standards. All funding applications or scientific manuscripts detailing newly generated antibodies should use non-animal-derived materials from naïve universal or engineered library sources that are defined at the sequence level. Grant requests and scientific reports proposing the use of antibodies generated through animal immunization should be challenged.
Academic institutions are urged to coordinate efforts to establish non-animal-derived libraries and broader access to services that will support research activities. The report signposts those manufacturers already leading the way in offering non-animal-derived antibodies in their catalogs.
The message is clear: through the harnessing of non-animal-based affinity products and technologies, we can make significant advances in safeguarding the welfare of millions of animals, while raising scientific standards concerning the use of antibody products in research, diagnostics, and therapeutics.
HuCAL and CysDisplay are trademarks of MorphoSys. Images copyright Bio-Rad Laboratories.
Amanda Turner ([email protected]) is product manager for custom antibody products at Bio-Rad Laboratories.
References
- Barroso J, Halder M, Whelan M. EURL ECVAM Recommendation on Non-Animal-Derived Antibodies. EUR 30185 EN, Publications Office of the European Union, Luxembourg; 2020.
non-animal-derived-antibodies. Accessed June 2020. - Bradbury A, Plückthun A. Reproducibility: Standardize antibodies used in research. Nature 2015; 518: 27–29.
- Knappik A, Ge L, Honegger A, et al. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J. Mol. Biol. 2000; 296: 57–86.
- Gray AC, Sidhu SS, Chandrasekera PC, et al.
Animal-Friendly Affinity Reagents: Replacing the Needless in the Haystack. Trends Biotechnol. 2016; 34(12): 960–969. - Rothe C, Urlinger S, Löhning C, et al. The Human Combinatorial Antibody Library HuCAL GOLD Combines Diversification of All Six CDRs According to the Natural Immune System with a Novel Display Method for Efficient Selection of High-Affinity Antibodies. J. Mol. Biol. 2008; 376(4): 1182–1200.
- Hatlem D, Trunk T, Linke D, Leo JC. Catching a SPY: Using the SpyCatcher-SpyTag and Related Systems for Labeling and Localizing Bacterial Proteins. Int. J. Mol. Sci. 2019; 20(9): 2129.

