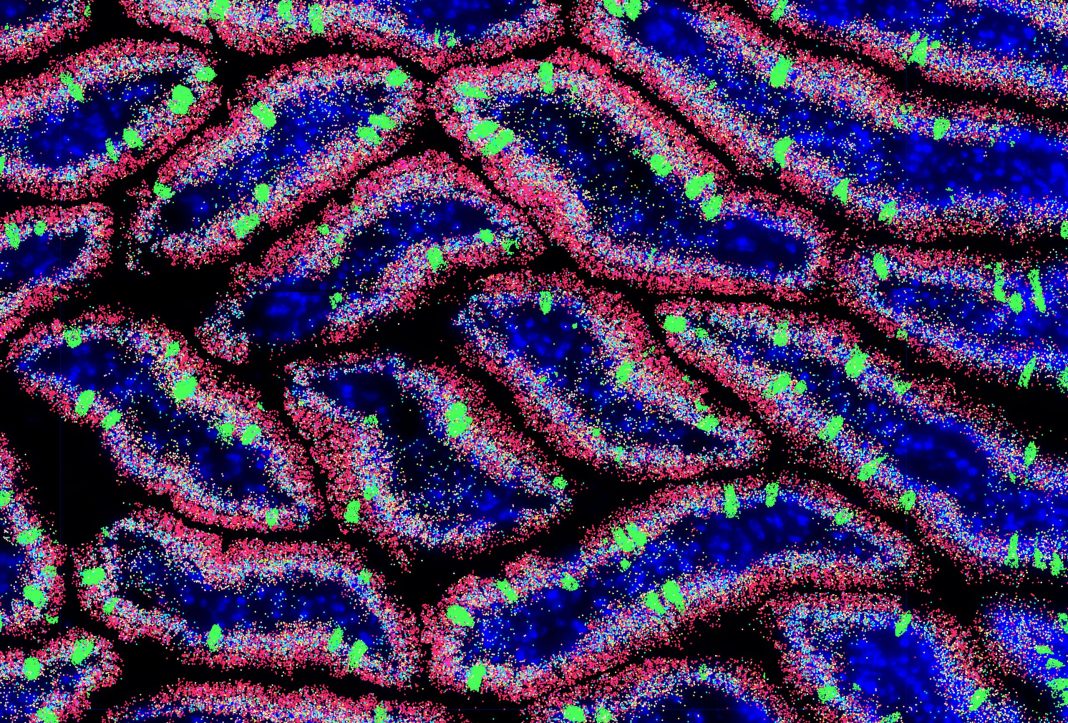
Spatial biology is poised to become the “next big thing” in omics. On the strength of new technologies, it promises to elevate an ambitious concept—the localization of tissue components at the cellular and molecular levels. Although the concept has been around for decades, spatial biology technologies are taking it to new heights. Indeed, these technologies promise to usher omics into a bright new era.
In a recent Nature Methods review entitled “Museum of Spatial Transcriptomics” (2022; 19: 534–546), Lior Pachter, PhD, professor of computational biology at California Institute of Technology, and his graduate student Lambda Moses, traced the origin of modern spatial techniques back to 1969. At that time, a method for the radioactive in situ hybridization of ribosomal RNA debuted. Since then, varied spatial methods has emerged.
Today, several techniques are in development for the analysis of proteins or RNA transcripts in the spatial context of tissues. And the commercialization of these techniques is being pursued by more than a dozen companies, including 10x Genomics, Akoya Biosciences, Lunaphore Technologies, NanoString Technologies, Rebus Biosystems, Resolve Biosciences, Veranome Biosystems, and Vizgen. These companies are releasing instruments and offering their own predictions and promises about what their platforms can do.

Stanford University
“It’s early days—very early days,” says Michael Snyder, PhD, professor of genetics at Stanford University. But the potential is huge. Understanding organization at the single-cell level is the next big wave.
It is clear that more researchers are adding spatial experiments to their research papers and budgets, and that more companies are pushing spatial technologies. Accordingly, GEN believes it is important to hear from the scientists at the spatial frontier. In this article, GEN shares what these scientists have to say about where spatial biology is now—and where it might be going.
Possibilities and practicalities
“We use lots of fancy terms, but spatial still means sectioning a piece of tissue and putting it on a slide,” notes Jasmine Plummer, PhD, founding director of the Center for Spatial Omics at St. Jude Children’s Hospital. She adds that while “spatial omics” is regarded as something entirely new, it is only the “omics” part that is new. The “spatial” part has been around since Ramón y Cajal was doing histology, that is, since the late 1800s.

Where is spatial today? Right now, Plummer says, it is still “old school.” Many scientists conduct spatial research the same way they have for decades—using antibody labeling techniques. The next phase of spatial will be determined by whether the newer, omics-driven spatial technologies are successful.
For the anatomists and histologists who use conventional antibody labeling, the transition to spatial omics entails greater costs. These investigators already have a cost-effective means of determining whether a patient has cancer or inflammatory bowel disease. They can resort to using an H&E stain on a slice of tissue—a procedure that costs about $5.00. If spatial omics were as economical, it would be adopted with little hesitation. “There is,” Plummer says, “not a single researcher who wouldn’t want [to use spatial omics] to look at the 20 proteins that they’ve been looking at individually for their entire lives, where all 20 are in the same space and [can be assessed simultaneously].” But alas, costs matter.
Players besides companies
At the Advances in Genome Biology and Technology (AGBT) meeting in June, attendees couldn’t turn around without bumping into a spatial company talking about its platforms. A talk given by Fei Chen, PhD, assistant professor of stem cell and regenerative biology at Harvard University and a core institute member at the Broad Institute, was a refreshing change. Although his slides were packed with spatial data, they were missing any references to commercial systems. That is because Chen has developed his own spatial technology—Slide-seq. Chen admits that he “doesn’t pay that much attention to what the companies are saying.”
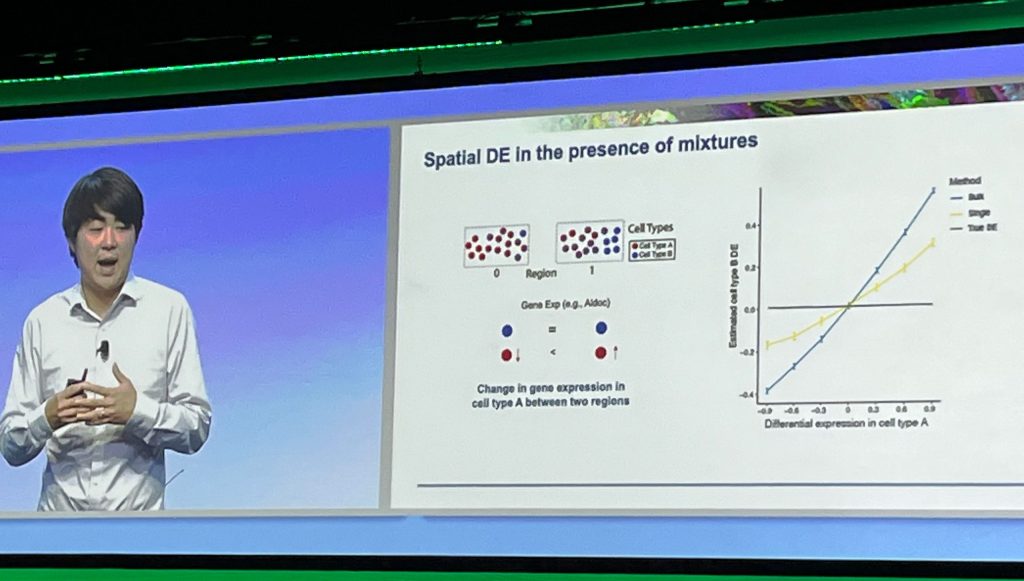
Why is spatial so powerful? According to Chen, spatial provides the ability to understand how cell-type compositions are changing within a context better than single cell analysis. Spatial is expected to be especially valuable as a means of characterizing cellular interactions and improving our understanding of intercellular communications at the gene level.
Single-cell analysis has been a powerful technology for discovering cell-type compositional changes, cell-state changes, and connecting phenotypes to genes. But it’s obvious that single-cell studies don’t put those findings in the context of tissues. And that is why people are excited about spatial. And many other companies have picked up on that excitement. Spatial has become a hot area of commercial innovation—much more so than single-cell analysis.
Where spatial is valuable
“A fibroblast is not a fibroblast is not a fibroblast,” Snyder declares. Each fibroblast, the veteran geneticist explains, has a profile that depends on the cell’s location and what the cell is next to. He thinks that uncovering cellular interactions will be “super cool,” especially since doing so promises to reveal how cells communicate with each other. Analyzing how cells are perturbed in disease states—like a viral infection—is also going to be interesting using spatial. Snyder says that researchers will be able to uncover biology that nobody has ever been able to get at before.
The Slide-seq data Chen showed at AGBT enabled an analysis of the gene expression changes that occurred in immune cells interacting with tumors. More specifically, the data allowed T cells and T-cell receptors (TCRs) to be examined in the context of tumors. Chen’s team was able to see the receptor, profile gene expression, and establish spatial context. Moreover, the team deconvoluted which factors were cell intrinsic, cell extrinsic, or dependent on the tumor context. The team is starting to uncover cell-to-cell interactions between tumor cells and the environment.
Chen believes that the ability to tease apart the interactions and the communication networks will be useful not just in cancer research, but in other fields such as stem cell biology and neurodegenerative disease. Spatial, he suggests, is impressive not just for the depth of its analyses, but for the breadth of its applications.
Scientific rigor needed
According to Plummer, the spatial field has reached the “I can do it” phase, where some researchers are satisfied to say, “Look at my pretty picture.” She admits that the field is not yet at the “I want to make a clinical biomarker” phase.
Does Snyder think that people are using spatial because it’s the new cool thing to do? Or are people asking important questions? He answers: “Yes and yes.” Snyder says that researchers must do spatial to stay current. If they don’t, manuscript and grant reviewers will ask why they don’t have spatial data. That’s the way science works; there is a lot of herding behavior.
Plummer says that spatial will likely follow a trajectory similar to the ones followed by next-generation sequencing (NGS) and single-cell analysis. New technologies allow people to be more successful in obtaining funding and publications. There is an incentive for researchers to adopt new technologies. That shift, she says, has not happened yet in spatial. She adds, however, that “we are primed as a research community to make the leap in spatial.”
Snyder agrees. There are “a ton of people” that have already jumped on the single-cell dissociated-cell bandwagon, he says. Those folks, he predicts, will just move over to spatial.
Chen thinks hard about how to use spatial, giving great consideration to experimental and analytical procedures. But he notes that the technologies in the spatial field have, in a general sense, outpaced this type of rigorous thinking. Nonetheless, Chen says spatial can he very useful for doing hypothesis generation or discovery, when applied appropriately.
At present, many people are preoccupied with validating their single-cell data. That, Chen says, is “probably not the right way to think about the problem.” He suggests that if the framework and the computational tools, are in place to support thinking about the problems the right way, spatial will be powerful for hypothesis generation.
This kind of spatial requires a lot of data—more data than spatial has been able to process to date. But this problem may be resolved through commercial development. More people are thinking about how to direct spatial to the right sorts of questions and conduct meaningful analyses. Plummer agrees that what we need are more numbers. The problem with spatial is that its signal-to-noise ratios are too low. But it’s important to get the data out there so that the analysis methods can improve.
Encouragingly, spatial doesn’t arouse skepticism of the sort that initially greeted single-cell technology. Already, the need for spatial is widely recognized. Spatial has an intuitive appeal because researchers have been doing histology and pathology for a long time. And so, spatial may become routine even more quickly than did single-cell technology.
Chen often receives collaboration requests—more than he has the bandwidth to handle. To expedite his evaluation of these requests, he asks potential collaborators to define the question that they want to answer, and to relate how they intend to frame the question in terms of spatial analysis.
Essentially, he tries to ensure that collaborators have the computational background to process (and think about how to work with) the data. Such a background is needed to support spatial experimentation.
Plummer adds that spatial is primed for the application of machine learning and artificial intelligence (AI) technologies. “We don’t even have to wait,” she says. Colleagues are already using these technologies to analyze information about tumor slices. She adds, however, that these technologies will need prodigious amounts of data if they are to conclude, for example, that “this is something different than this one.”
An extra constituency
Thus far, this article has emphasized similarities between the early days of single-cell technology and the current status of spatial technology. But Plummer notes a distinction between the two. Whereas single-cell technology is dominated by a single constituency—people who used to rely on bulk RNA sequencing—spatial may have two constituencies. The first consists of people who transitioned from bulk RNA sequencing to single-cell technology and who are now interested in transitioning to spatial. The second consists of people who are interested in pathology.
This extra constituency includes molecular anatomists who work with mouse models to learn more about their favorite genes. They have a few antibodies that they’ve been using over a long career. Even if they’ve never done a single-cell experiment, they care about their tissues. A single-cell experiment might not answer their question, but spatial might prove attractive to them.
There are many researchers who are not into sequencing and omics who are used to looking at tissues. Many investigators are still diagnosing diseases in hospitals using tissue sections. You don’t have to be an omics person to adopt spatial technology. That said, spatial is much more cost prohibitive to these researchers. It’s not as daunting to scientists who are already spending thousands of dollars on single-cell experiments.
All in the name of progress
Chen asks us to think back to the 1960s and consider how much thought scientists had to put into planning their experiments. He offers the example set by the Nobel laureates François Jacob and Jacques Monod. These scientists had to painstakingly marshal what were, by today’s standards, scarce and feeble resources before they could even think of running the experiments that ultimately revealed a fundamental paradigm of molecular biology.
Today, science is different. It enjoys abundance and power. There are more scientists, more technologies, and more experiments. Does the profligacy result in experiments that are, individually, less monumental than those of decades past? Probably, answers Chen.
Today, few scientists devote enormous forethought to their experiments. But the upside is that there is greater progress being made than in the sixties. “I don’t think that that’s the problem,” Chen remarks. Instead, the problem, he suggests, is that the ability to collect the data has outpaced the ability to analyze it and to frame problems the right way. But that’s not a bad problem to have, he says. It’s just something we have to solve as a field.