Reproduction is typically how humans share genetic information. This genetic transfer is referred to as vertical or passing traits “down,” so to speak.
However, some organisms share genes through horizontal DNA exchange, meaning a transfer that is not parent-to-offspring. This process is well understood in simple organisms, such as bacteria, where it plays a key role in their ability to exchange the genes that encode for antibiotic resistance. A new paper investigates the possible role of horizontal transfer in mammals, by analyzing the presence of transposons (or “jumping genes”) in different organisms’ genomes.
The article, “Horizontal transfer of BovB and L1 retrotransposons in eukaryotes,” is published in Genome Biology. The researchers from the University of Adelaide, led by David Adelson, Ph.D., professor at the University of Adelaide, department of genetics and evolution, found that horizontal gene transfer was much more widespread than was previously thought.
Transposable elements occupy roughly half of the human genome. Some, such as the long interspersed element (LINE) retrotransposons, move to a new site after copying themselves while leaving an old version behind, leading to an amplified number in the genome. Two of the most abundant retrotransposon families in eukaryotes are LINE-1 (L1) and Bovine-B (BovB.)
In 2013, Dr. Adelson showed that horizontal transfer of BovB was significantly more widespread than believed and demonstrated a possible mechanism of transfer via two plausible arthropod vectors, specifically reptile ticks. This work was published in the Proceedings of the National Academy of Sciences of the United States of America.
Regarding their ongoing work on this subject, Dr. Adelson says, “Jumping genes, properly called retrotransposons, copy and paste themselves around genomes, and in genomes of other species. How they do this is not yet known although insects like ticks or mosquitoes or possibly viruses may be involved – it's still a big puzzle.”
Dr. Adelson and colleagues hypothesized that the presence of the L1 retrotransposon in mammals is due to an ancient horizontal transfer event. In order to study this, they extracted millions of retrotransposon sequences from a dataset consisting of 759 plant, fungal, and animal genomes.
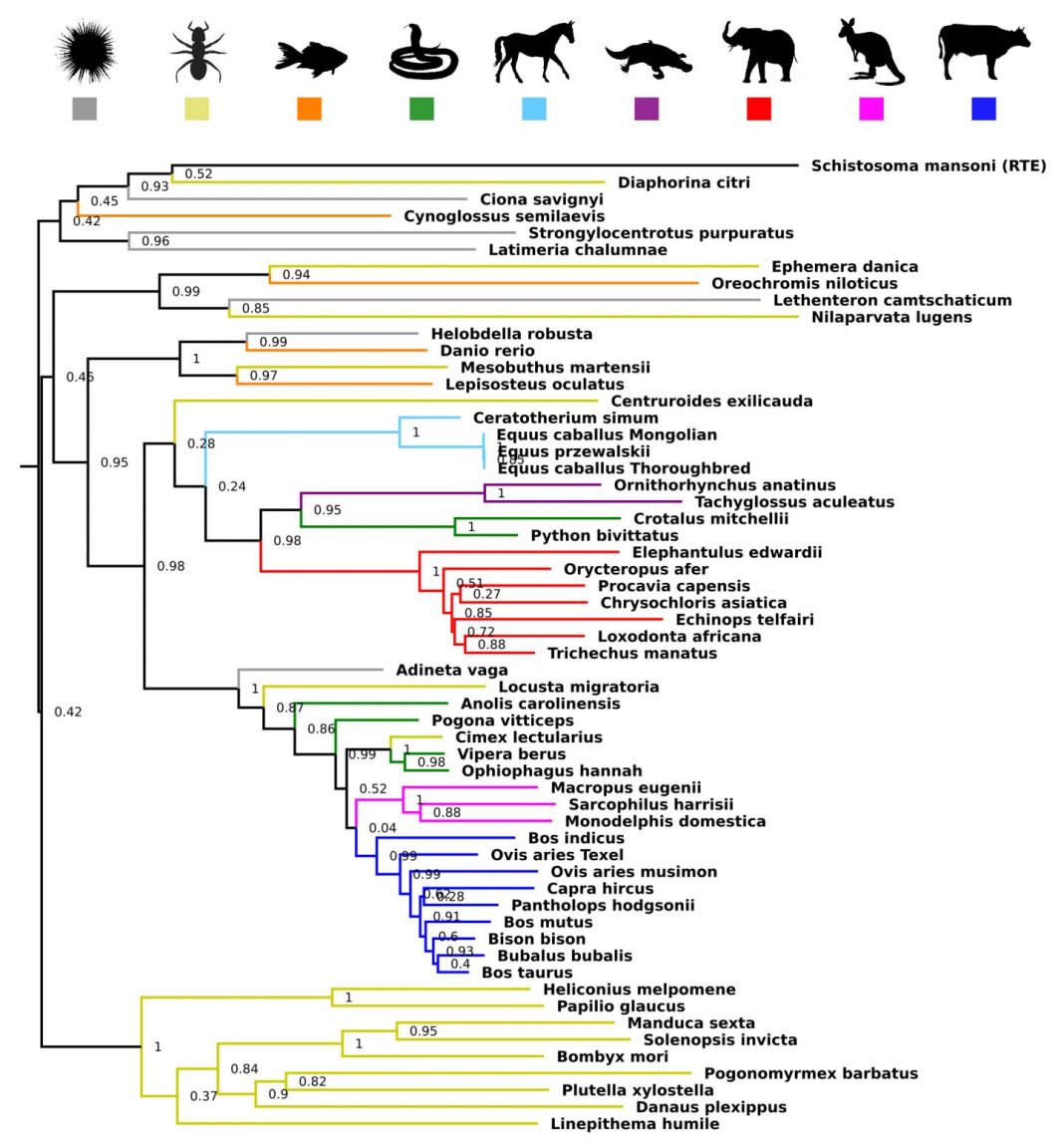
In the current work, the researchers illustrated that both the L1 and BovB retrotransposons are present in a diverse array of species including mammals, reptiles, fish, amphibians, arthropods, and primitive species such as sea urchins and sea squirts and have similar distributions among the organisms.
They found that the BovB elements were more widespread than previously thought and both had a patchy distribution among the genomes analyzed in this study. The authors were particularly curious about their results in monotremes (egg-laying mammals such as the duck-billed platypus) as they were the only species to contain BovBs and lack L1s.
The researchers used three criteria to detect the presence of a horizontal transfer event of a transposable element. The transposable elements should have a patchy distribution among organisms, very high sequence similarity despite being in dissimilar organisms and inconsistencies between transposon element phylogenetic tree topology and species relationships.
Interestingly, they found that cross-species transfers, even between plants and animals, have occurred frequently throughout evolution and identified multiple possible L1 horizontal transfer events in eukaryotic species, primarily involving Tx-like L1s in marine eukaryotes. They also found new parasite vectors of horizontal transfer such as the bed bug, leech, and locust and discovered BovB occurrences in new lineages such as bat and frog.
The authors write that “We therefore speculate that the speciation of therian mammals was driven in part by the effect of L1 retrotransposition on genome structure and function, including regulatory effects on transcriptional networks.”
Due to the large amount of the mammalian genome that is comprised of transposable elements, these results support the role of horizontal transfer creating long-term genomic change in new host organisms. The authors write that “With the advancement of genome sequencing technologies and bioinformatics tools, we anticipate our study to be a valuable resource for inferring horizontal transfer from large-scale genomic data.”