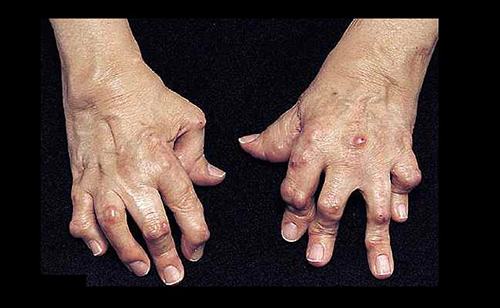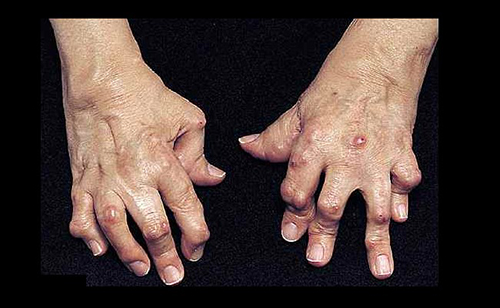
In rheumatoid arthritis (RA), gene expression varies from joint to joint, causing the disease to progress along different molecular pathways, each of which may respond to different drugs. The finding that RA varies by location—affecting some joints early, others late, and some not at all—helps puts old observations in perspective. It also points to new possibilities for precision medicine.
The finding helps explain, in part, why drugs treating RA vary in effect—why, for example, a treatment that might work in arthritic knees isn't effective in an arthritic hip. Also, the finding suggests that arthritic processes in different joints may call for targeted treatments.
Although past studies had suggested that pathogenesis of RA is similar in all affected joints, scientists based at the University of California, San Diego (UCSD) must have sensed that something was amiss in the state of RA research. So, to set things right, these scientists put aside any self-doubt, and they resolved to probe the cellular processes and molecular pathways underlying RA. In particular, they took a close look at the DNA methylation and transcriptome signatures of fibroblast-like synoviocytes (FLSs), specialized cells that line the inside of joints.
The scientists, who were led by Wei Wang, Ph.D., and Gary S. Firestein, M.D., found signatures that not only discriminated RA FLSs from osteoarthritis FLSs, but also distinguished RA FLSs isolated from knees and hips. Details of this work appeared June 10 in the journal Nature Communications, in an article entitled, “Joint-Specific DNA Methylation and Transcriptome Signatures in Rheumatoid Arthritis Identify Distinct Pathogenic Processes.”
“Using genome-wide methods, we show differences between RA knee and hip FLSs in the methylation of genes encoding biological pathways, such as IL-6 signalling via JAK-STAT pathway,” wrote the article’s authors. “Furthermore, differentially expressed genes are identified between knee and hip FLSs using RNA-sequencing.”
While osteoarthritis tends to localize in weight-bearing joints where cartilage is specifically worn away, RA is distributed more symmetrically—both hands may be affected equally, for example—and often evolves from the small joints of the hands and wrists to the larger weight-bearing joints. These observations suggested to the UCSD scientists that there was more to RA than was previously dreamt of.
“We hypothesized that changes in epigenetic modifications and gene expression between FLSs in different joints might potentially contribute to differences in synovial inflammation and responses to clinical treatment,” said Dr. Wang.
To their own selves true, the researchers undertook a study that may have looked a little crazy to their peers, and they eventually discovered that DNA methylation—a fundamental, life-long process in which a methyl group is added or removed from the cytosine molecule in DNA to promote or suppress gene activity and expression—does in fact vary between FLS from the knees and hips of RA patients.
“We showed that the epigenetic marks vary from joint to joint in diseases like RA,” explained Dr. Firestein. “Even more importantly, the differences involved key genes and pathways that are designed to be blocked by new RA treatments. This might provide an explanation as to why some joints improve while others do not, even though they are exposed to the same drug.”
The work, added Dr. Firestein, “opens up the potential for precision medicine approaches that allow us to target all of the joints, not just a subset. It has broad implications for how we evaluate new drugs in clinical trials as well.”