June 15, 2012 (Vol. 32, No. 12)
Facilitating Multisite Bioprocess Transfer: Multi-Instrument and Multiplatform Stability
The biopharmaceutical industry has grown exponentially and more companies are now operating in a global market with sites thousands of miles apart. The need is now greater than ever for robust bioprocess monitoring solutions that can provide consistent instrument-to-instrument results.
The seamless transfer of information across multiple sites relies heavily on the comparability of process data from various technologies, ensuring effective monitoring and control of critical process parameters. The goal of this study was to assess the comparability of Nova Biomedical’s BioProfile® chemistry and gas analyzers across several development and manufacturing sites in the U.S.
In addition, the long-term performance stability of the BioProfile systems was also tested. Five BioProfile FLEX and four BioProfile 100 Plus analyzers were used to determine linearity, precision, accuracy, and instrument-to-instrument comparability.
The age of the instruments used for this study ranged from new to over eight-years old, with instruments that had tested several hundred samples to over 20,000 samples on a given analyzer.
Facility Growth
The use of small-scale bioreactors in development and commercial manufacturing facilities has grown from just a few to over several hundred instruments, with manufacturing capacities in the hundreds of thousands of liters. It is nearly impossible to utilize the same analytical devices in manufacturing suites that were used in the development of a process.
Scientists and engineers must feel confident that comparable results can be achieved between multiple analytical systems across parallel development labs and up through commercial manufacturing suites. Furthermore, manufacturing campaigns are run months to years after a process has been developed, requiring robust analytical technologies that scientists and engineers can rely on.
This study was performed in partnership with a large biopharmaceutical firm. This cross-site correlation study was initiated with the goal of obtaining robustness data, linearity and precision data, and ultimately to test the interchangeability of equipment.
Quality Control Verification
For this study, a single lot of QC material for each level was used. The expected range (the range within which verifies proper system function) for each parameter and each level was programmed into the BioProfile FLEX and the BioProfile 100 Plus analyzers. All levels of QC material were equilibrated to room temperature prior to analysis.
Additional QC material provided for the measurement of particles for the BioProfile FLEX Cell Density/Viability module. This material contains polystyrene beads and was used to determine proper fluid flow, dilution, and calibration of the CDV modules and to confirm proper function of the imaging system on all FLEX analyzers used in this study.
For this study, it was important to use samples of known concentration, when available. For pH, gas, electrolyte, nutrient/metabolite, and osmolality measurements, Nova’s QC systems were used. These controls are manufactured to provide standards of known concentration across the analytical range.
Control levels 1, 2, and 3 provide for the measurement of pH, pCO2, pO2, glucose, lactate, NH4+, Na+, K+, Ca++, and osmolality. Levels 4 and 5 provide for the measurement of glucose, lactate, glutamine, and glutamate. To provide a greater sample set and additional points along the linear range of the analyzers, various standards were diluted with water for injection.
The expected values for the additional levels were determined mathematically by using the dilution ratio used to create the additional levels. It should be noted, however, that the additional levels created were not used for pH or gas measurements as these would have equilibrated with room air and would not provide acceptable measurements.
Cell culture samples obtained from a bioreactor were used for viable cell density, total cell density, and viability measurements.
Unless otherwise noted, all sample sets were analyzed in triplicate on each analyzer.
Calculations were used to determine the mean, standard deviation, and coefficient of variation for the data generated in this study.
Results
As we were looking for instrument-to-instrument agreement, the data in this study show results compared to a reference FLEX analyzer. This analyzer was arbitrarily chosen among the group of instruments used in this study. It is identified as FLEX 1.
For measured gasses (pH, pO2, pCO2), linearity was strong with correlation coefficients of greater than 0.98 (graphs not shown). In addition, the instrument exhibited a high level of comparability with a standard deviation of 0.03 pH units and a coefficient of variability of 0.4%. For pO2 and pCO2, standard deviation was 2.29 and 7.94 mmHg and coefficient of variability was 5.2% and 7.7%, respectively.
Figure 1 shows the linearity profiles for glucose, lactate, glutamine, and glutamate. For glucose and lactate, linearity was very strong with correlation coefficients of greater than 0.99 for both. In addition, the instrument exhibited a high level of comparability with a standard deviation of 0.11 and 0.06 g/L and a coefficient of variability of 4.9% and 2.9% for glucose and lactate, respectively.
Similarly, correlation coefficients were greater than 0.97 for both glutamine and glutamate. In addition, the instrument exhibited a high level of comparability with a standard deviation of 0.13 and 0.07 mmol/L and a coefficient of variability of 6.0% and 5.2% for glutamine and glutamate, respectively.
It should be noted that the linearity and variability for glutamine are not as strong as the other parameters tested. Though the samples were controlled in the best possible manner (samples stored on ice prior to analysis), the instability of glutamine often results in higher variability in large experiments such as this.
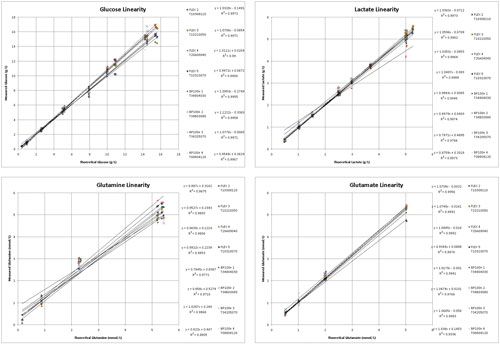
Figure 1. Glucose, lactate, glutamine, and glutamate linearity profiles
Figure 2 illustrates the linearity profiles for ammonium NH4+, Na+, and K+. NH4+, Na+, and K+ also exhibited very strong linearity correlation coefficients of greater than 0.99 for all three analytes. Standard deviation was 0.09, 2.23 and 0.09 mmol/L and coefficient of variability was 5.3%, 2.5% and 2.8% for NH4+, Na+ and K+, respectively.
Figure 2 also includes the total cell density linearity profile for the BioProfile FLEX analyzers. In the figure, results obtained on the BioProfile CDV module are compared to results obtained on the Cedex cell counter, the reference instrument used in this laboratory.
The data was generated from bioreactor cultures seeded at varying densities, providing a range of total cell density and viability samples.
Due to intellectual property concerns, the viability data is not shown. However, a range of cultures were analyzed with viabilities ranging from approximately 45% to 99% viability. BioProfile FLEX viabilities were within 3% compared to the Cedex analyzers.
Nova Biomedical provides published specifications for all parameters (excluding CDV) as measured on a single analyzer for both within run and day-to-day precision. This study incorporated nine BioProfile instruments, with samples analyzed over the course of several weeks.
The results reported in this study demonstrate that not only can one expect a single BioProfile instrument to meet published specifications for precision, but multiple instruments can also fall well within the published specifications of a single instrument. This data further supports the BioProfile analyzers as extremely robust analytical tools for cell culture development and manufacturing process monitoring.
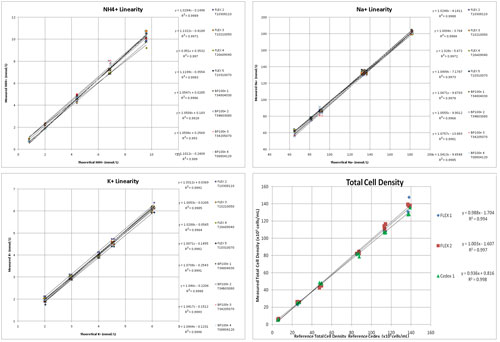
Figure 2. NH4+, Na+, K+, and total cell density linearity profiles
Matthew McRae ([email protected]) is worldwide technical applications manager, John McHale is vp, quality assurance and technical support, Scott Granger is senior technical applications specialist, Brian Goulart is technical applications and training specialist, and Elizabeth Kilcoyne is international BioProfile support manager at Nova Biomedical.



