Genome editing technologies, such as CRISPR, are being applied to better understand basic biological systems as well as to research new kinds of gene and cell therapies. The CRISPR-Cas9 system comprises a Cas9 endonuclease protein that forms a complex with a guide RNA (gRNA) molecule, which then localizes to a target DNA sequence where the gene editing takes place. The CRISPR protein complex specifically recognizes the DNA target because the target sequence complements the gRNA and contains a protospacer adjacent motif (PAM) necessary for compatibility with the Cas9 enzyme. This powerful new technology for genetic manipulation and engineering is being investigatedfor various potential therapeutic applications.1
With new technologies, however, come new unknowns, including the degree to which phenomena, such as off-target editing (OTE) may have adverse biological effects. Although we currently have no standard baseline or reference ranges for detection of OTE levels in clinical applications, that does not mean that there is no benefit to measuring OTE levels or to establishing such standards.
Although the precision of CRISPR has been greatly increased with the development of improved tools such as high-fidelity CRISPR-Cas nucleases that elicit less OTE than their wild-type counterparts, CRISPR is still liable to edit at unintended sites in the genome. Therefore, regulatory authorities require the demonstration of OTE assessments in the development of new clinical applications that utilize CRISPR. Also, the measurement of OTE levels is often a necessary step in experimental workflows, for example, to direct the selection of gRNAs, a critical component of the CRISPR targeting system.
Several methods to assess CRISPR OTE sites have been created. These methods generally include two key stages. First, the potential OTE sites are identified; second, these nominated OTE sites are sequenced after CRISPR editing to investigate the amount of editing occurring at each of the sites. There are multiple tools available for each of the two stages.
Nominating OTE sites
For identifying the OTE sites, there are mainly two approaches used to nominate the locations of potential OTE. One type relies on computational algorithms that predict potential OTE sites based on sequence homology between the target genome and the CRISPR targeting components, namely the Cas enzyme PAM and the gRNA spacer sequences (Figure 1). However, these OTE assessments in silico have been shown to overestimate the OTE sites.2
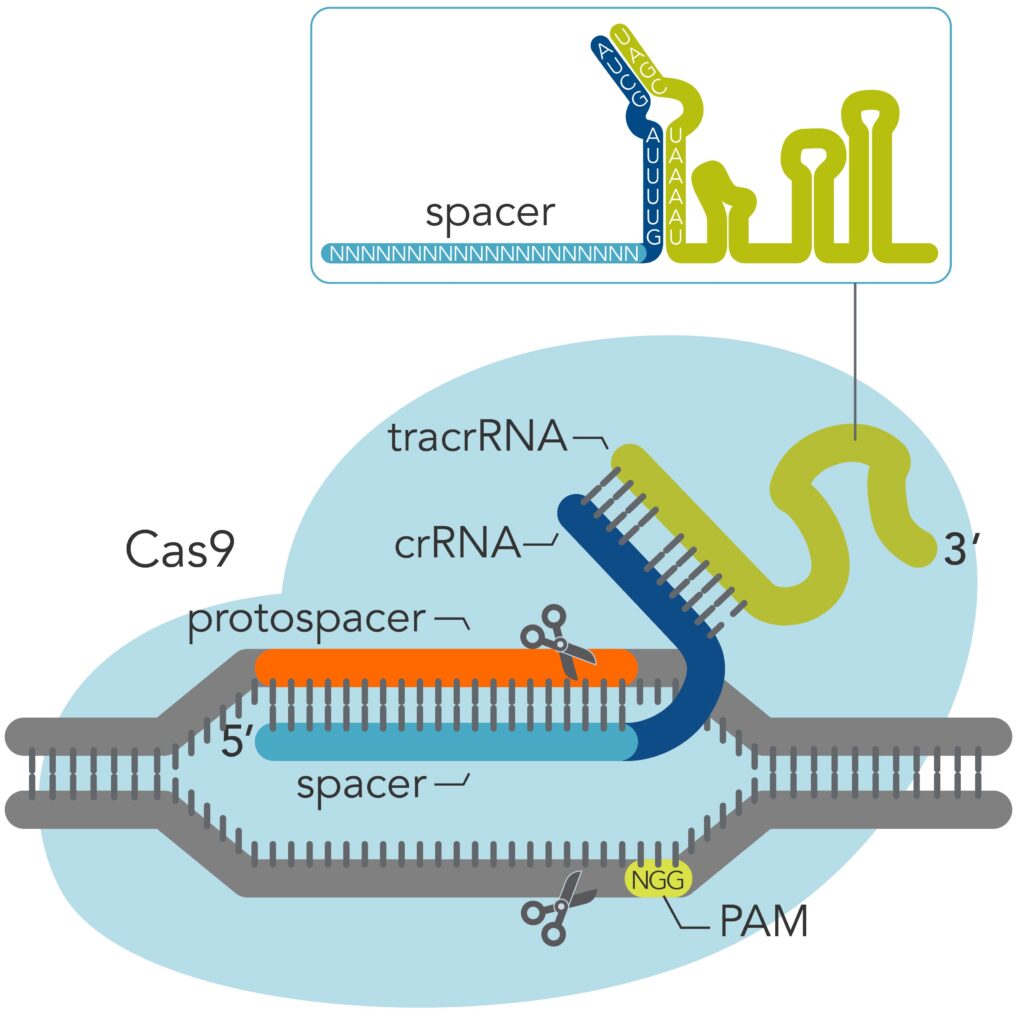
The other type applies an empirical approach that measures OTEsites using a combination of genome-wide assays with next-generation sequencing (NGS). Typically, one in silico tool and one empirical technique both are applied to comply with regulatory guidance regarding the use of two orthogonal methods in identifying the OTE sites. The computational tools employ artificial intelligence (AI) algorithms to nominate where the OTE may occur, based mainly on predicting where the CRISPR complex can bind. Many of these tools have evolved from methods originally designed to predict sites for on-target editing. The deep learning achieved with AI has enabled methods such as DeepCRISPR and CNN_std to predict OTE sites with a potential reduction in false positives.
These methods, however, are based on identifying matches between the CRISPR complex and target genome. Consequently, there is a tendency toward overestimation. Moreover, these methods require that the sequence of the target genome be completely annotated. Due to these limitations, empirical methods are also required to obtain a complementary assessment of the OTE sites.
Empirical methods are assays that aim to capture CRISPR gene editing events. These assays can be cell-based or biochemical. An example of a cell-based assay is the Genome-wide Unbiased Identification of Double-stranded breaks Evaluated by Sequencing (GUIDE-seq).3
The GUIDE-seq assay is a research tool that is applicable when a standard model system is used. GUIDE-seq employs a standard human embryonic kidney (HEK) cell line and a specially modified gRNA. The stable HEK293 cells constitutively express the Cas9 nuclease, meaning that the Cas9 protein is continuously being produced in these cells. This greatly increases the chances of OTE happening.
To increase these chances even further, a gRNA is used that has been heavily chemically modified to be very stable. This means that the gRNA will persist in the cells for a longer time before being degraded and therefore, provide more time for OTE to occur. Generating as much OTE as possible is key because that information enables further research into the OTE sites of interest.
Detecting OTE activity
Once the OTE sites have been identified, the potential amount of editing taking place at each site can then be quantified. This can be performed in the model system as well as in primary cells that have undergone CRISPR gene editing. At Integrated DNA Technologies
(IDT), the rhAmpSeq™ CRISPR Analysis System* is applied to quantify OTE activity.
The rhAmpSeq CRISPR Analysis System is performed in multiplex and is therefore relatively rapid and robust, while enabling the specificidentification of editing events resulting from CRISPR-mediated double- stranded breaks in genomic DNA.4 It does this using proprietary rhAmp PCR technology based on a two-step PCR process.
The first PCR step amplifies targeted sections of the genome very specifically through the mitigation of primer dimer. This is achieved by using blocked primers that contain RNA bases, and the ribonuclease enzyme, RNase H2, which cleaves only DNA:RNA complexes
(Figure 2). The amplicons from the first step are then amplified further in the second PCR step, using standard indexing primers.
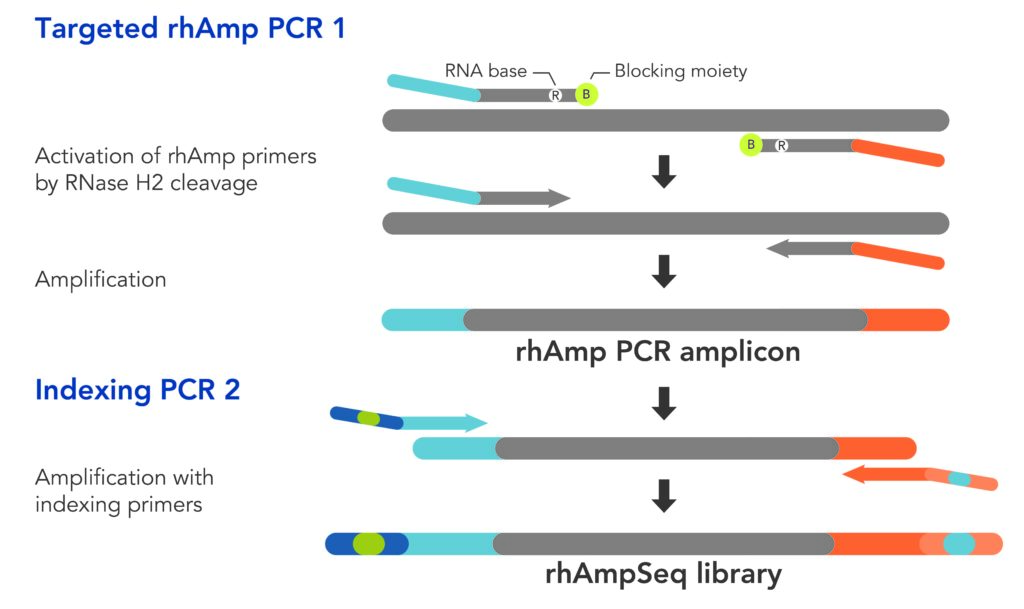
The resulting amplicon library can then be sequenced using NGS, and the CRISPR-Cas edits can be quantified. The CRISPR gene edits of the genome—both on- and off-target—can also be characterized as insertions or deletions.
Measuring and minimizing OTE when developing clinical applications
GUIDE-seq and the rhAmpSeq CRISPR Analysis System have recently been used in the research of Katy Rezvani, MD, PhD, professor of stem cell transplantation and cellular therapy at the University of Texas MD Anderson Cancer Center. Rezvani is pioneering the research of CRISPR gene editing to modify human immune cells—such as T cells and natural killer (NK) cells—to better recognize and attack tumor cells.
Rezvani does this by introducing a gene into the cells that results in their expressing a chimeric antigen receptor (CAR). This research has shown that the CAR proteins are able to bind to complementary proteins on tumor cells, thereby enhancing and redirecting the immune system to attack the cancer.
In some of her latest published research,5 Rezvani and her team took human NK cells from umbilical cord blood, genetically engineered them with a fourth-generation “armored” CAR to create CAR-NK cells, then used CRISPR to knockout the CISH gene in the CAR-NK cells. The CISH gene encodes for the cytokine-inducible Src homology 2–containing (CIS) protein, a checkpoint regulator of NK cell antitumor activity. By knocking out the CISH gene, Rezvani and her team were able to enhance the cytotoxic activity of the CAR-NK cells against cancer in human cell and animal models.
On- and off-target editing was assessed as part of the development process of the CRISPR system used. GUIDE-seq and the rhAmpSeq CRISPR Analysis System were applied to measure OTE for the selection of the optimal Cas9 nuclease and gRNA. Less OTE was observed after switching to a high-fidelity Cas9 nuclease, Alt-R™ HiFi Cas9. Various gRNA candidates were screened, and the gRNA that induced the least amount of OTE was similarly chosen.
Once the CRISPR system was developed and the experiments performed, GUIDE-seq was used in a model system to investigate OTE. Subsequently, the rhAmpSeq CRISPR Analysis System was used to quantify the amount of OTE occurring in the CAR-NK genome at the sites found by GUIDE-seq. The CAR-NK OTE sites and events measured were minimal, largely due to the use of the highly efficient and specific Cas9 enzyme in a ribonucleoprotein complex with the optimal gRNA. Thus, Rezvani is using CRISPR for the gene editing of cells to improve the targeting and effectiveness of potential cancer therapies.
Ayal Hendel, PhD, a senior lecturer and principal investigator at Bar-Ilan University in Israel, is leading a team researching CRISPR-Cas9 genome editing as a tool to eventually enable the development of a curative therapy for genetic diseases, such as severe combined immunodeficiency (SCID)—widely known as the “bubble boy” disease. They are also using Alt-R HiFi Cas9 in RNP complexes in their R&D experiments and employing GUIDE-seq and rhAmpSeq to ensure OTE events are minimized.6
Another researcher conducting studies for the advancement of CRISPR therapeutics is Matthew Porteus, MD, PhD, professor at the Stanford Center for Definitive and Curative Medicine. He and his team are focusing on genetic diseases that affect children, including sickle-cell disease and β-thalassemia. Their research has informed early-stage clinical trials where the safety profile and proof-of-concept are currently being evaluated.
Many biomedical researchers anticipate that CRISPR gene editing will next be investigated for in vivo therapies, that is, gene editing of cells within the body as opposed to ex vivo, or editing of cells outside of a patient’s body. It is hoped that by that time the biomedical and clinical research community will have a much better understanding of both on- and off target gene editing, including the nascent establishment of reference standards and acceptable detection limits for OTE. These will certainly be critical for helping to make CRISPR-based therapies and cures a reality.
References
1. Ledford H. Quest to use CRISPR against disease gains ground. Nature 2020; 577(7789):
156. DOI: doi: 10.1038/d41586-019-03919-0.
2. Chaudhari HG, Penterman J, Whitton HJ, et al. Evaluation of Homology-Independent
CRISPR-Cas9 Off-Target Assessment Methods. CRISPR J. 2020; 3(6): 440–453. DOI: 10.1089/
crispr.2020.0053.
3. Tsai SQ, Zheng Z, Nguyen NT, et al. GUIDE-seq enables genome-wide profiling of off-target
cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015; 33(2): 187–197. DOI: 10.1038/
nbt.3117.
4. Schubert M, Rettig GR, Kurgan G, et al. Evaluate CRISPR-Cas9 edits quickly and accurately
with rhAmpSeq targeted sequencing. Application note. Integrated DNA Technologies.
https://go.idtdna.com/rhAmpSeqCRISPR.html.
5. Daher M, Basar R, Gokdemir E, et al. Targeting a cytokine checkpoint enhances the
fitness of armored cord blood CAR-NK cells. Blood 2021; 137(5): 624–636. DOI: 10.1182/
blood.2020007748.
6. Shapiro J, Iancu O, Jacobi AM, et al. Increasing CRISPR Efficiency and Measuring Its Specificity
in HSPCs Using a Clinically Relevant System. Mol. Ther. Methods Clin. Dev. 2020; 17:
1097-1107. DOI: 10.1016/j.omtm.2020.04.027.
Rolf Turk, PhD, is a senior staff scientist at Integrated DNA Technologies.
Website: www.idtdna.com.
* The IDT products identified in this article are for research use only and not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations.
