May 15, 2006 (Vol. 26, No. 10)
Detecting Low-Abundance Biomarkers without Immunoaffinity Depletion of Abundant Proteins
Blood is a rich and readily accessible source for the detection of diagnostic markers and therapeutic targets in many human diseases, such as cardiovascular diseases, neurological disorders, and cancer. Currently, however, only a handful of plasma proteins are routinely used in the clinic for diagnostic purposes. This is largely due to the lack of an effective technology platform that rapidly detects and quantifies specific changes of the low-abundance proteins in plasma. The dynamic range of plasma protein concentrations comprises greater than ten orders of magnitude, and the low-abundance proteins are difficult to detect as they are often masked by the high-abundance proteins, particularly albumin.
Immunoaffinity chromatography has been used to remove the most abundant plasma proteins, yielding an enriched pool of low-abundance proteins for studies. Although this strategy has achieved some success, it has drawbacks.
Applied Biomics (www.appliedbiomics.com) has developed a technology by which thousands of low-abundance plasma proteins can be resolved and quantitatively analyzed on 2-D gels without immunoaffinity depletion of the abundant proteins. This technology allows for more sensitive detection and accurate quantification of low-abundance plasma proteins.
Analysis of the proteome of plasma and serum represents a challenge due to the wide range of protein concentrations and high structural complexity of the constituent proteins in these samples. In serum and plasma, the quantities of high-abundance proteins and some low-abundanceproteins span over 10 orders of magnitude. The 12 most abundant plasma proteins comprise more than 90% of total protein content in plasma.
Although changes in some resident proteins have proven to be of predictive value in several human diseases, it is generally established that the low-abundance proteins likely contain most of the useful biomarkers. To date, the most common approach is immunoaffinity-based depletion. Although this strategy has increased the number of detectable proteins in plasma/serum analysis, it has several major drawbacks.
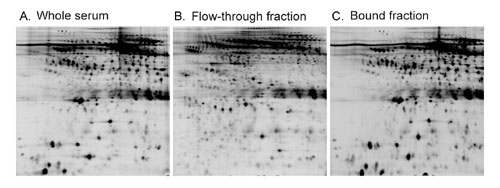
First, up to 90% of potential protein biomarkers are associated with the highly abundant carrier proteins in blood. Thus, depletion of the high-abundance proteins also removes many potentially important marker proteins. In addition, some low-abundance proteins may be retained in the column through nonspecific binding to antibodies and/or beads, resulting in their loss from the flow-through fractions. This has been shown by many studies and is also demonstrated by our experiment (Figure 1).
Second, extensive sample handling during the depletion process increases the chance of sample loss, protein degradation, and modification artifacts, resulting in substantial sample-to-sample variation.
Third, samples are diluted during the depletion process, making them less suitable for many downstream analyses. For many applications, steps must be taken to concentrate the proteins, which again can result in the loss of certain proteins and introduce analysis deviation.
Finally, the depletion process is time-consuming, and the immunoaffinity columns are rather expensive. Regeneration of the columns inevitably results in the decline of antigen binding efficiency, which further increases the cost and contributes to sample-to-sample variation.
Fig.1: Immunoaffinity depletion of the high-abundance proteins removes many potentially important marker proteins. In addition to the 12 proteins that are designed to bind to the column, a large number of low-abundance proteins are also bound to the column
Profiling without Immunoaffinity Depletion
Applied Biomics’ strategy focuses on the low-abundance, low molecular weight (LMW) proteins in blood. In general, only the LMW proteins and peptides are small enough to leak from the surrounding tissues into blood to produce diagnostic traces. Furthermore, many signaling molecules transmitted through the bloodstream are small proteins, often below 40 kDa. The LMW portion of the blood proteome is a treasure trove of diagnostic information.
In order to specifically detect LMW protein biomarkers in blood and compare their relative abundance between disease samples and normal controls, Applied Biomics optimized 2-D gel analysis by adopting three technology improvements. First, plasma/serum samples are denatured under conditions that are designed to effectively solubilize small proteins and peptides, but not the larger proteins that are over 45 kDa. Under such conditions, the LMW proteins can be efficiently focused in the isoelectric focusing (IEF) strips (GE Healthcare). High molecular weight proteins and high-abundance proteins (most of which are greater than 45 kDa) are poorly absorbed into the IEF strips.
Furthermore, high percentage (>13.5%) SDS-PAGE gels are used for the second-dimension separation, thereby maximizing resolution of the LMW proteins. This has resulted in a significantly increased number of detectable proteins at the LMW range of the plasma/serum proteome (Figure 2). Because the samples are thoroughly denatured, LMW proteins are completely dissociated from the carrier proteins so that no loss of LMW proteins will result from the treatment.
Second, 2-D differential in-gel electrophoresis (2-D DIGE), rather than regular 2-D gel electrophoresis, is used for the detection and quantification of protein differences between test and control samples. Each sample is separately labeled with a distinct fluorescent CyDye (Cy2, Cy3, or Cy5). The labeled samples are mixed together and resolved on a single 2-D gel, and the corresponding protein spot patterns are visualized by multichannel scanning of the same gel. The resulting images are analyzed both visually and by using software such as DeCyder (GE Healthcare).
Fluorescent labeling increases the detection sensitivity over standard colorimetric staining methods. More protein spots can be detected and analyzed: using the optimized protocol for LMW plasma proteins, Coomassie brilliant blue staining typically has a detection limit in the range of 300 protein spots on a 13×15 cm gel, silver staining approximately 1,200, whereas fluorescent labeling with CyDyes typically allows for the detection of more than 2,000 protein spots.
Post-electrophoretic staining with fluorescent dyes (such as SYPRO Ruby) displays a similar detection sensitivity to that of CyDye-labeling. However, post-electrophoretic processing increases the possibility of protein losses particularly in the LMW range.
Most critical is the increased confidence with 2-D DIGE that differences in spot fluorescence intensity are purely attributable to biological and not experimental variation, as realized by the complete elimination of gel-to-gel variability through using a single gel for the comparison of different plasma samples. Because every protein spot has its own internal control, small differences in protein abundance (as low as 15%) between samples can be detected with greater than 95% statistical confidence.
Third, using 2-D DIGE technology allows several plasma samples (each labeled with a different fluorescent CyDye) to be mixed and partitioned simultaneously. For example, centrifugation of the mixture in a spin-style concentrator with a defined molecular weight cut-off size can proportionally remove high molecular weight proteins from each sample. The co-partitioning process is intrinsically more reliable than partitioning in parallel, as the latter will inevitably introduce sample-to-sample variability during the partitioning steps and compromise quantitative accuracy.
Figure 3 shows an example that demonstrates the potential of these strategies for biomarker discovery. It is now possible to analyze the plasma proteome from multiple species in a potentially rapid and large-scale capacity for biomarker discovery, drug target discovery, and toxicology studies.
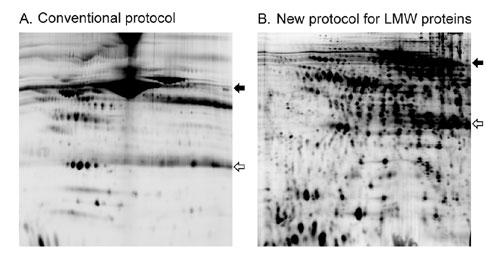
Fig.2: Focusing on the LMW plasma proteins. Typically, more than 2,000 protein spots can be resolved using Applied Biomics’ conditiions.
Broad Applications
2-D gel electrophoresis is one of the most popular and effective discovery tools in proteomics research. To date, however, its application in studying the plasma/serum proteome is limited, as are other available proteomics technologies. The massive complexity of the proteins in plasma and serum presents a tremendous challenge and calls for a highly parallel display and quantification strategy.
Applied Biomics’ 2-D DIGE-based technology allows parallel protein analysis on 2-D gels and holds promise to accelerate the discovery of novel protein biomarkers in plasma and serum. Furthermore, the co-partitioning process described here is applicable to many other sample types, such as urine, saliva, cell, and tissue protein extracts, to minimize sample-to-sample variation and facilitate quantitative analysis.
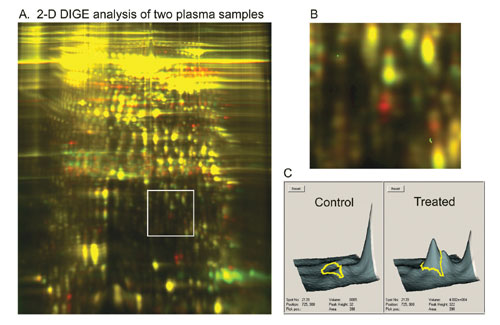
Fig.3: Identifying differentially expressed protein markers in plasma samples. The 3-D images show the two protein spots that are clearly overexpressed in the treated plasma sample.