University of Tokyo researchers and colleagues at the RIKEN Research Institute have compiled a first-of-its-kind genetic database for autoimmune and autoinflammatory diseases. It’s hoped that the resource will offer up new insights into how immune disorders develop, and potentially aid in drug discovery. Scientists also hope that the atlas of immune-related genome data may eventually be applied to investigations of infectious diseases, such as COVID-19.
“To understand diseases, a deep comprehension of the function of genetic variants is essential,” said University of Tokyo project research associate Mineto Ota, MD, PhD, a clinical rheumatologist and expert in functional genomics. “With this data set, we can connect the data about changes to DNA sequence associated with a disease to genes and cell types that are important for disease pathogenesis.” Ota is lead author of the team’s published study in Cell, which is titled, “Dynamic landscape of immune cell-specific gene regulation in immune-mediated diseases.” The project was carried out in collaboration with scientists at Chugai Pharmaceutical.
Immune-mediated diseases (IMDs) comprise a wide range of pathologies, from autoimmunity to autoinflammation, the authors wrote. And while aberrant inflammatory cytokine activity in IMDs indicates altered functions of immune cell, “ … little is known about the responsible genes or even relevant cell types underlying IMDs,” they noted.
Genome-wide association studies (GWAS) have revealed a number of genomic loci that are associated with IMDs. But many of the genetic variants identified in such association studies are located in noncoding regions of DNA that regulate the “on” or “off” expression of genes, and not in the gene-coding sequences. “GWAS have revealed a number of genomic loci that are associated with complex traits and diseases, including IMD,” the team noted. “The majority of these variants lie in noncoding genomic regions, especially regulatory elements such as enhancers and promoters.”
To uncover the function of regulatory DNA, a different type of genome analysis, called expression quantitative trait loci (eQTL), aims to connect differences in DNA sequence to differences in gene expression. With eQTL data, researchers can make more informed guesses about the purpose of regulatory DNA sequences, how variants in the regulatory sequence might affect expression of the genes it regulates and how those differences in gene expression cause disease.
Other immune-focused eQTL studies have been performed, but prior research efforts included only healthy volunteers and examined a limited number of cell types. “… studies of immune cells from healthy donors might not capture the range of eQTL effects found in disease states,” the investigators wrote. “Also, while experimental stimulation of immune cells can mimic disease status to some extent, it does not always reflect the in vivo physiologic environment.”
Ota added, “Inflammatory conditions create very different physical characteristics in immune cells compared to the same cells in a healthy condition. The genetic variants associated with immune conditions may only function in the diseased state, so for this type of genetic study, it was very important to get samples from real patients.”
For their newly reported studies, the research team sequenced the full genomes of 79 healthy volunteers and 337 patients diagnosed with any of 10 different categories of immune-mediated diseases, including rheumatoid arthritis, systemic lupus erythematosus, and systemic sclerosis. All volunteers were of Japanese ancestry.
Understanding immune-mediated diseases is challenging because although each disease is clinically distinct, there are many overlaps and patients with the same diagnosis may show very different symptoms. “Due to all this diversity, there’s a limit to how much you can learn studying one immune-mediated disease at a time,” Ota added. “But if we study 10 diseases together, it gives a bigger picture about these types of diseases.”
After completing full genome sequencing, researchers isolated 28 different types of immune-related cells from volunteers’ blood samples and measured gene expression in those cells. “We see that each immune cell type has distinctive eQTL results in the atlas, which can tell us how gene regulation differs between cell types and exactly which cell type is important for developing which disease,” said Ota.
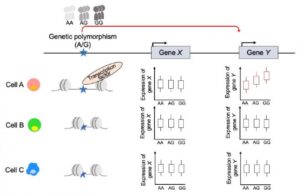
The authors reported, “In gene-expression analysis, our comparison of multiple IMDs in a single platform permitted us to characterize each disease (patient) and identify genes that were associated specifically with a single disease, as well as pathways that were dysregulated in a group of IMDs … Our dataset enabled us to detect physiological (disease) context-driven perturbations of eQTL effects in each specific cell type.”
The researchers have named the resulting resource the Immune Cell Gene Expression Atlas from the University of Tokyo (ImmuNexUT), and say it represents the largest eQTL data set built using volunteers of East Asian ancestry. “We identified immune cell type-specific eQTL effects, including previously unreported rare cell types, their diversity under physiological inflammatory conditions, and significant association of these eQTLs with IMD genetics,” they concluded.
“In this field so far, large-scale genomic and functional genomic studies have been mainly conducted with European donors, although population diversity is critical for precise understanding of the genomic functions,” Ota stated. “This eQTL atlas of Japanese subjects is also meaningful for overcoming this European-centric bias and further investigating the functions of DNA variants in combination with European data sets.”
Researchers hope the ImmuNexUT database will continue to grow and eventually lead to better outcomes for patients. “Our dataset has distinct and advantageous characteristics compared to previously reported resources, including comprehensive immune cell subdivision, variation of immunological conditions among donors, and homogeneous non-European donors,” they said.