April 1, 2014 (Vol. 34, No. 7)
Software Pipeline for Gene Candidate Identification
The past decade has seen accelerated discovery of the causal genes underlying Mendelian and complex diseases as well as progress toward the goal of personalized medicine.
Technological achievements including the human genome sequence, the collection of millions of marker single nucleotide polymorphisms (SNPs), and microarrays have allowed the transition from single-gene studies to genotyping whole genomes.
Microarray-based genotyping of large numbers of case and control individuals enabled genome-wide association studies (GWAS) whereby SNPs associated with a disease could be identified statistically. This approach has identified numerous loci that contribute to complex diseases and traits.
However, the common SNPs identified by microarray-based GWAS are rarely causal, but instead are linked to the genes and/or variants of interest. Moreover, the loci identified typically combine to contribute a fraction of the observed heritability for the disease under study.
It is increasingly evident that the “missing heritability” involves rare (minor allele frequency
The advent of next-generation sequencing (NGS) offers a cost-effective solution to these problems by making it possible to determine the complete sequence of all gene regions (the exome) or even the whole genome of an individual. By mapping NGS data to the human genome reference sequence, common, rare, and private variants in that individual can be identified.
Further, variants in coding regions can be prioritized based on their functional impact. Exome sequencing of even small numbers of patients has identified causal genes for a number of dominant and recessive Mendelian diseases. Now, exome sequencing of much larger cohorts is also beginning to elucidate the causes of complex diseases.
Computational Challenges
The number of whole-exome, not to mention whole-genome, NGS datasets that are needed to conduct these studies pose significant computational challenges. First, the massive amount of raw data (typically 10–30 GB for a single exome and 300–400 GB for a whole genome) requires substantial computer resources for processing as well as for storage and management.
Second, there is a series of computational tools required (Figure 1): (1) a large capacity NGS reference-guided assembler, (2) a variation detection module, (3) a variant annotation module(s), (4) a visualization package for inspecting alignments and variants calls, and (5) an analytics module for comparing variants across samples including statistical analyses and discrete filtering.
For all but the most sophisticated clinical research groups, stringing together and running the disparate software tools needed to accomplish these tasks is an insurmountable hurdle.
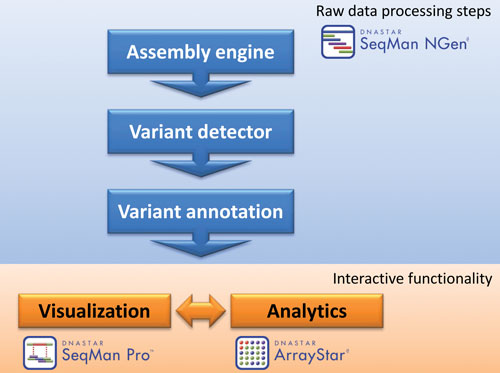
Figure 1. A series of computational tools is required to process whole-exome and whole-genome datasets. The job of stringing these tools together is technically demanding.
Integrated Software Pipeline
The Lasergene Genomics Suite (DNASTAR) is a commercial-grade, integrated software pipeline that facilitates data flow with an easy-to-use, intuitive, graphical interface. The suite consists of three programs: SeqMan NGen, SeqMan Pro, and ArrayStar.
SeqMan NGen has a step-by-step wizard for project setup and data management. Specified data is then fed into a proprietary nonmemory bound assembly engine for aligning NGS datasets of any size to the human genome and producing fully gapped alignments. Those alignments are evaluated by a SNP and small indel detection module that uses a Bayesian probability model to call bases and genotypes at each position.
Finally, variants are annotated with their impact on the corresponding coding region(s), the GERP++ score of that position, and whether it is present in either the dbSNP or COSMIC databases. BAM-formatted alignment, variant call and annotation files of each chromosome are packaged together as output. SeqMan NGen can assemble deep human exome datasets in under two hours and whole genomes in less than a day on modestly priced (
Assembly packages from a disease study can be loaded either individually into SeqMan Pro for visual inspection of the alignments and variant calls or together into ArrayStar for association analyses using statistical and discrete filtering options as outlined below.
Case Study: Kabuki Syndrome
As a demonstration of DNASTAR’s pipeline, exome datasets from the published Kabuki syndrome study (Ng et. al., Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 42, 30-35 (2010)) were obtained through dbGaP. Kabuki syndrome is a rare Mendelian disorder (approximately 400 cases reported worldwide) caused by autosomal dominant mutations.
The ten case and eight control exome datasets were independently aligned to the human genome reference sequence using SeqMan NGen, which also identified and annotated variants. Variants from each assembly were then loaded together into ArrayStar resulting in 5,729,442 independent positions located in 32,024 genes across all samples after coalescing. The samples were then organized into two groups, Kabuki and control, to facilitate subsequent discrete filtering.
We first filtered at the variant level (Figure 2) making three assumptions based on knowledge of the disease: (1) causal mutations would be non-synonymous changes, (2) causal mutations arose de novo so variants would occur in only one case sample, and (3) no control sample would have any of the mutations. Stringent quality metric thresholds were also imposed to reduce noise. 11,366 variants in 6,352 genes met the criteria and were saved as a “SNP set.”
The case-only SNP set was then used as the variant pool in a second filtering step (Figure 2). This time to identify genes with mutations that met the following criteria: (1) mutations were inactivating (nonsense or frameshift), (2) they were rare and therefore not in dbSNP and (3) they were dominant and therefore occurred as heterozygotes. 845 genes met those criteria in at least one case sample. However, by increasing the level of detectance to 7 of 10 case samples the number of candidates was reduced to one, MLL2, consistent with the results of Ng et. al.
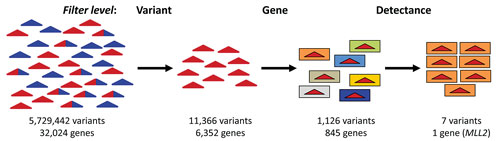
Figure 2. Discrete filtering series to identify causal gene for Kabuki syndrome. Colored triangles indicate variants from cases (red), controls (blue), or both (red/blue). Colored boxes indicate different genes.
Conclusions
Exome sequencing has revolutionized our ability to detect common, rare, and private variants in the coding genes of an individual. By sequencing case and control cohorts and then comparing across the spectrum of variants, the genetic causes of Mendelian and complex diseases are being uncovered. NGS technologies and facile software pipelines that integrate assembly, variant calling/annotation and association analyses are essential partners in this endeavor.
The recent arrival of the $1,000 whole genome promises to expand the discovery of critical variants throughout the genome. Software pipelines that keep pace with data gathering and analytical advances will be crucial for expanding the search into the causes of human disease.
The continuing expansion of DNASTAR’s Genomics Suite in support of clinical sequencing, from gene panels to exome and whole genome association studies, is in recognition of the revolutionary impact this field will have on human health.
Tim Durfee, Ph.D. ([email protected]), is a principal scientist, Dan Nash is a senior software engineer, Schuyler Baldwin is a senior software engineer, Kerri Phillips is a product manager, and Frederick R. Blattner, Ph.D., is founder, president, and CEO of DNASTAR.



