Researchers across multiple disciplines are using metabolomics—the systematic identification and quantification of small-molecule metabolic products—for an expanding array of applications. This trend is accelerating progress in areas such as translational medicine, drug development, and food quality analysis.
Broadening the base of metabolomics users are the suppliers of metabolomics systems. Besides elaborating instrumentation platforms around key technologies such as mass spectrometry, these suppliers are refining associated workflows and software.
This work isn’t being performed in a supplier bubble. Indeed, several suppliers are partnering with research institutions to help metabolomics technology become more user-friendly, even as it gains sophistication and power.
Grappling with a systemic disease
Bruker recently indicated that it has partnered with the Australian National Phenome Centre (ANPC) to support a new “frontline response to combat the COVID-19 threat.” Researchers at the ANPC are using instrumentation from Bruker to help them perform metabolic analyses on samples taken from COVID-19 patients and controls.
“The metabolic effects of SARS-CoV-2 infection on human blood plasma were characterized using multiplatform metabolic phenotyping with nuclear magnetic resonance (NMR) spectroscopy and liquid chromatography–mass spectrometry (LC-MS),” ANPC researchers reported in a recent study.1 “[We constructed] an exceptionally strong hybrid NMR-MS model that enabled detailed metabolic discrimination between the groups and their biochemical relationships.”
By probing the profound metabolic alterations that accompany COVID-19, the researchers identified markers “distinctive of a multisystem involvement” and “consistent with the reported extensive microvascular effects that would be expected to compromise multiple organ functions.”
Going forward, the researchers hope to build models that will predict variation in the severity of the disease, help clinicians understand differential responses to therapeutic interventions, and address other uncertainties. “One unanswered question about COVID-19,” noted Lucy Woods, PhD, business unit manager, Phenomics & Metabolomics, Bruker, “is why sufferers experience such a wide range of symptoms.”
Woods adds that metabolomics researchers must obtain reproducible measurements across assays and acquire an accurate picture of the presence of metabolites. Although extensive analyte libraries are available, a large proportion of analytes remain unidentified, necessitating the use of orthogonal techniques.
According to Bruker, ANPC researchers will use several of the company’s systems, including the Avance IVDr NMR platform and the Impact II, timsTOF Pro, and Solarix MS platforms, as well as various data modeling approaches, to perform metabolic analyses of the molecular, physical, and biochemical characteristics of blood plasma and urine samples to create informative translational models. These models, Bruker asserts, will predict variation in the severity of the disease and help clinicians understand differential responses to therapeutic interventions.
Coping with growing sample numbers
As more researchers explore the use of metabolomics and sample numbers increase, there is a need to enhance throughput without sacrificing coverage, identification, and accessibility. Many metabolites or lipids can exist in different isomeric forms, some biologically active and others not. They need to be properly identified if their biological significance is to be understood.
To facilitate discovery work, Waters offers high-resolution mass spectrometers, the Synapt XS high-resolution mass spectrometer and the Select Series Cyclic IMS system, both enabled with ion mobility separation technology that provides orthogonal structural information for identification. Ion mobility increases coverage, condenses chromatographic run times, and captures the sample complexity.2
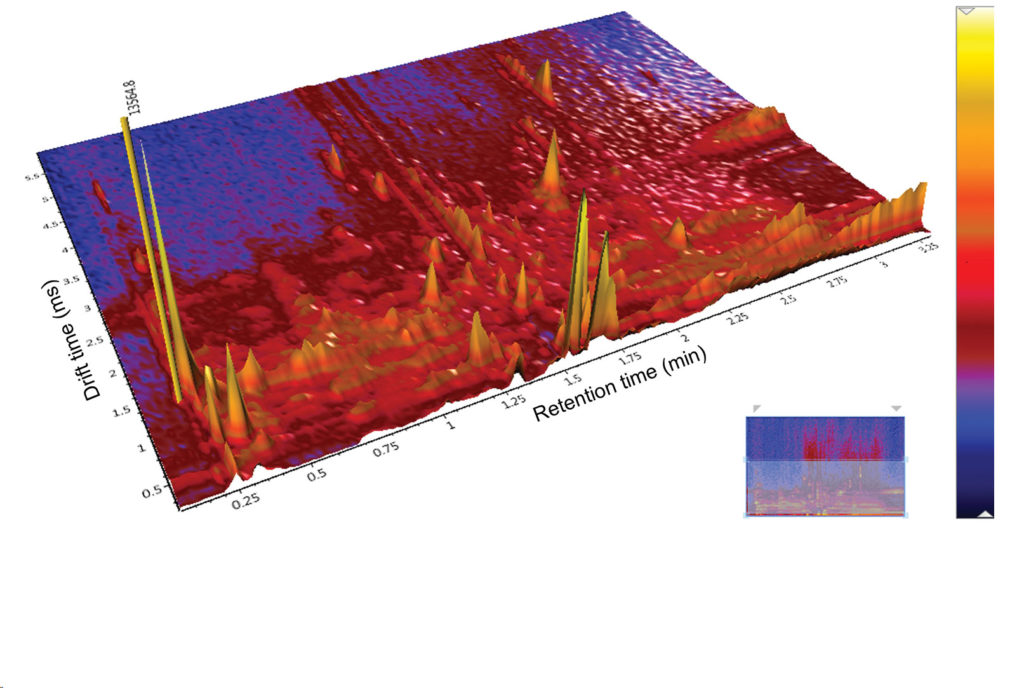
In a metabolomics study that processes, for example, 1,000 samples, “ultraperformance liquid chromatography (UPLC) ion mobility MS can decrease analysis time from 1,300 to 200 hours,” says Suraj Dhungana, PhD, global biomedical research market segment manager, Waters. “For structural characterization of metabolites, our Cyclic IMS ion mobility mass spectrometer provides unique, on-demand variable ion mobility resolution. When isomeric metabolites cannot be separated using liquid chromatography, we can separate ions using ion mobility based on their rotationally averaged collision cross section (CCS).”
By exploiting its “racetrack” geometry, the Cyclic IMS system can improve ion mobility resolution as needed to separate small differences in CCS values by increasing the number of times the ions cycle around a mobility cell—the racetrack. This allows separation and very clean fragment ion spectrums for library matching.
To address the identification challenge for the natural products research community, Waters has partnered with a research group led by Roger G. Linington, PhD, at Simon Fraser University. This group maintains the Natural Products Atlas, a database of microbially derived natural product structures. The searchable version of the database contains 25,000 microbial compounds that can be directly queried with Waters discovery metabolomics software (Progenesis QI) and screening software (UNIFI).
“For added flexibility, we offer desorption electrospray ionization (DESI) and MALDI sources for discovery, targeted, and spatial studies,” adds Dhungana. “Metabolomics or lipidomics experiments and a DESI imaging experiment can be performed on the same instrument platform by simply swapping the source.”
Ensuring food quality and security
The food industry uses metabolomics to address the taste or smell of foods, develop new functional foods, and improve fermented food quality, says Eberhardt R. Kuhn, PhD, marketing manager, Food and Consumer Products, Shimadzu Scientific Instruments. Other applications include discovering patterns of abundance ratios of certain metabolites to confirm geographical origin of foods and beverages to verify food authenticity and prevent food fraud and identification of dietary biomarkers to screen for food allergens.
Shimadzu offers a variety of MS products for metabolomics research, such as triple-quadrupole LC-MS, quadrupole time-of-flight (Q-TOF) LC-MS, triple-quadrupole gas chromatography MS, and MALDI-TOF MS, as well as software platforms and databases that simplify workflows.
“Because it has a footprint the size of a sheet of paper, the MALDImini-1 Digital Ion Trap mass spectrometer can be placed on a workbench for a more convenient, efficient workflow,” asserts Kuhn. “The system’s digital ion trap uses rectangular wave RF to allow ion trapping up to 70,000 Da. Furthermore, the tandem MS (MS/MS) and three-stage MS (MS3) functionality allows comprehensive structural analysis of unknown metabolite biomolecules.”
When analyzing LC-MS/MS data, the procedure known as peak picking is a huge bottleneck requiring visual confirmation. Equipped with algorithms developed using artificial intelligence, Shimadzu’s Peakintelligence software automatically detects peaks appearing in chromatograms, substantially reducing the need for visual confirmation.
Shimadzu’s Metabolites Method Package Suite is a compilation of seven existing products including LC-MS/MS method packages, multiple reaction monitoring (MRM) libraries, and a GC-MS metabolite database. It enables the analysis of more than 1,900 metabolites without needing to investigate separation conditions, MRM optimization, or parameter settings. Metabolites span both hydrophilic and hydrophobic compounds.
The Metabolites Method Package Suite also incorporates the Multi-omics Analysis Package, which supports both regular and large-volume data analysis and interpretation, and contains metabolic pathways and other contour maps, making it easy to visualize fluctuations in the quantitative values of metabolites across metabolic pathways. Data filtering functions and statistical analysis can be applied to the network of compound relationships.
Accommodating different levels of expertise
Metabolomics provides a comprehensive snapshot of small molecules in a biological system. In medicine, metabolomics aids in understanding disease progression at the biochemical level revealing disease metabolic signatures; in an industrial setting, it can be used to optimize the production of monoclonal antibodies.
A metabolomics pilot study may total roughly 50 samples, while a study consisting of multiple experimental groups can reach several hundreds, and population-based biobank and repository studies can surpass thousands. As study size increases, high-quality quantitative data must be generated consistently over extended periods.
“Since metabolomics can be used to answer a variety of scientific questions, new scientists are entering the field with little to no knowledge on the use of LC-MS. We are also seeing traditional LC-MS users who are not familiar with the analytical requirements and nuances trying their hand at metabolomics,” says Amanda Souza, metabolomics program manager, Thermo Fisher Scientific.
This year, the company launched two new instruments as part of its Orbitrap Exploris mass spectrometer portfolio, the Orbitrap Exploris 240 Mass Spectrometer and the Orbitrap Exploris 120 Mass Spectrometer. These systems offer an easy-to-use and intuitive user interface for the mass spectrometry novice, and they come with application-specific method templates for metabolomics and lipidomics experiments providing recommended parameter settings. Instrument calibration is streamlined and accomplished by clicking on a check box.
To provide valuable biological insights, robust reproducible measurements and confident metabolite identifications are required. Collaborative efforts show the Orbitrap Exploris 240 MS robustly generates reproducible measurements over multiple days with sub-ppm mass accuracy, consistent signal response, and long-term stability. In addition, AcquireX acquisition allows for more unique unknown compounds with fragmentation spectra for increased annotation confidence.
Lastly, Compound Discoverer software can support large-scale metabolomics data analysis across multiple sample batches while providing statistical analysis, annotation tools, and pathway mapping.
Balancing perfection and practicality
“Researchers are recognizing that complete unbiased coverage of the metabolome is not practical,” says Steven M. Fischer, technical market director, Academia and Government, Agilent Technologies. “In the early days of metabolomics, a lot of effort was made trying to find the ‘perfect’ extraction protocol. That has now given way to more pragmatic approaches, whereby analysts focus on subsets of the metabolome.
“The result is less breadth, but more certainty about what was detected and how much was present. Coverage and identification issues will become more challenging as researchers try to extend metabolomics to single-cell analysis and tissue imaging.”
To help researchers’ address issues such as compound coverage, identification, and quantification, Agilent develops various technologies. Many of Agilent’s technologies—gas chromatography (GC), liquid chromatography (LC), capillary electrophoresis (CE), and supercritical fluid chromatography (SFC)—can be used in metabolomics analysis after they have been coupled to a mass spectrometer, such as a single-quadrupole, triple-quadrupole, TOF, or Q-TOF instrument. Each mass spectrometer has its advantages and disadvantages.
In addition to instrumentation, Agilent develops separation columns and solid-phase extraction (SPE) kits for metabolomics. For example, Agilent’s hydrophilic interaction liquid chromatography column (HILIC-Z) is designed to provide highly reproducible results in analyses of polar metabolites by high-performance liquid chromatography (HPLC). An SPE material (Captiva EMR-lipid) developed by Agilent allows for selective removal of lipids from biological samples, and it helps tremendously with the challenge of sample preparation. Specialized software enables metabolomics analysis.
The range of problems that can be studied with metabolomics will continue to expand. In the near term, metabolomics will focus on developing standardized methods, says Fischer. Such methods, he adds, reflect a growing interest in data sharing that is due, in part, to an expanding community of metabolomics researchers.
References
1. Kimhofer T, Lodge S, Whiley L, et al. Integrative Modeling of Quantitative Plasma Lipoprotein, Metabolic, and Amino Acid Data Reveals a Multiorgan Pathological Signature of SARS-CoV‑2 Infection. J. Proteome Res. 2020; August 17: online. DOI: 10.1021/acs.jproteome.0c00519.
2. King AM, Mullin LG, Wilson ID, et al. Development of a Rapid Profiling Method for the Analysis of Polar Analytes in Urine Using HILIC–MS and Ion Mobility Enabled HILIC–MS. Metabolomics 2019; 15: 17. DOI: 10.1007/s11306-019-1474-9.



