Back in 2000, after the Human Genome Project’s leaders announced that a working draft of the human genome had been completed, GEN kicked off its coverage of the event by quoting from a New Yorker cartoon. “Whatever will we think about,” one cartoon figure asked another, “now that the genome project is almost complete?” Now, 20 years later, the quote seems more apposite than ever. With exquisite slyness, it suggests that we need to be careful about the word “complete.” It even leaves us thinking that what we call complete is really a beginning.
To better appreciate how the Human Genome Project’s first working draft was the start and not the end of something, let’s review a little history. And when we’re done—to the extent that any such endeavor can be “done”—we may appreciate another quote. This one doesn’t come from a cartoon caption, but a novel by William Faulkner. “The past is never dead,” he wrote. “It’s not even past.”
The beginning of the beginning
Our story starts with a secret meeting. It was held in May 2000 and organized by Ari Patrinos, PhD, then the director of the Office of Biological and Environmental Research in the U.S. Department of Energy. He had arranged to host Francis S. Collins, MD, PhD, the director of the National Human Genome Research Institute select NHGRI union and Craig Venter, PhD, the CEO of Celera Genomics. This meeting select and others) paved the way for an historic joint announcement on June 26—with President Bill Clinton at the White House—that the working draft of the Human Genome Project select HGP) was complete.
“We have reached a milestone that we promised to get to just about now, that is, covering the genome in what we call a working draft of the human sequence,” Collins declared. “That is not to say that we have it all finished and zipped up and every last letter precisely identified. That will take a number of additional steps and probably the better part of the next couple of years to achieve.” That proved a slightly optimistic timeline.
Whether it was the talented team of researchers, the furious competition with Celera that marked—and marred—the HGP’s final stages, or select as Collins joked) the beer and pizza Patrinos served at those secret meetings—the completion of the draft sequence was considered a momentous success, helped by the temporary ceasefire between leaders of the two rival teams select see the sidebar “Reading the Genome”).
“Today, we are learning the language in which God created life,” Clinton said, paraphrasing Galileo, who insisted that the natural philosophy, even the universe itself, was written in the language of mathematics. “[We] have caught the first glimpse of our own instruction book,” Clinton added. “It will revolutionize the diagnosis, prevention, and treatment of most, if not all, human diseases.”
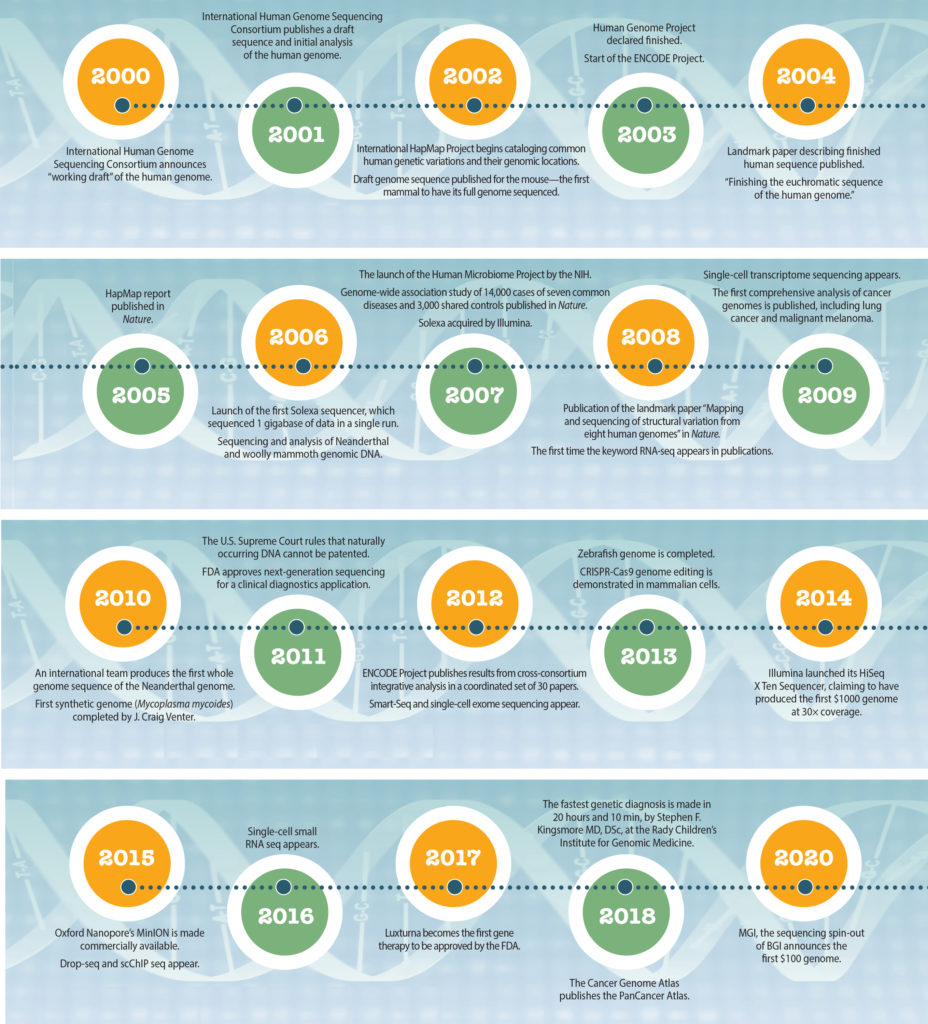
From passing glimpse to ceaseless scrutiny
Twenty years after the announcement, the HGP and the effort to create a pristine human reference genome has become intertwined with almost every aspect of biology and medicine. But where has it had the greatest influence?

Adam Felsenfeld, PhD
“It is hard to choose one,” remarks Adam Felsenfeld, PhD, NHGRI program director in the Division of Genome Sciences. “Having a human reference genome informs such a broad swath of human biology.”
At a high level, Felsenfeld says, the most significant result is “the ability to conceptualize the entire genome, or at least all the genes, as a finite number of things.” Adopting a comprehensive mindset—thinking genomically—has allowed scientists to formulate new questions and develop new technologies. These activities, he continues, have led to “all the genomics advances that we enjoy.” One such advance is the ability to assess all the variants underlying a particular genetic disease. Another is the ability to integrate the reference sequence with other data select including comparative genomics, gene expression, and other omics datasets). Yet another is the ability to relate genotype and phenotype.
One area of biology propelled forward by the HGP was the development of “faster and cheaper genome sequencing technology,” notes Jane Carlton, PhD, director of genomic sequencing at the New York University Center for Genomics and Systems Biology. Researchers learned that exciting biology could be gleaned from a whole genome sequence, and this realization sparked genome projects for a multitude of species. Carlton says that it has encouraged scientists to think, “If it has a genome, let’s sequence it.”
“Everybody is a genomicist because deep sequencing is an enabling technology,” she observes. “[It] has become so commonplace, it is no longer a limiting factor.” Genomics became all-pervasive, she asserts, because “the publication of the first human genome assembly and associated analyses showed what could be achieved.”

Indeed, whole fields have been catapulted forward due to the HGP. “[The HGP] is the underlying achievement behind what has been accomplished in the field of gene therapy research, and what we continue to work to achieve” notes Federico Mingozzi, PhD, chief scientific officer at Spark Therapeutics. The HGP, he continues, “provided the roadmap of genetic information required to bring the first gene therapy for a genetic disease [Luxturna] in the United States to patients.”
Contributions to other fields, such as synthetic biology, may be less obvious, but are no less important. Keith Robison, PhD, long-time genomics blogger at omicsomics.blogspot.com, tells GEN that in the synthetic biology field, “the technology spillover from the HGP has meant that we have the tools both to survey the biological world and to use sequencing to test our designs and as a readout.”
The advent of genomic medicine
One industry that could not exist without the HGP is personal genetics. Eric Green, MD, PhD, Collins’ successor as NHGRI director, tells GEN that he “did not expect to see genomic medicine become a reality during my career, perhaps not even in my lifetime.” But he notes that one of the advances of the HGP is “the initial realization of meaningful applications of genomics to the practice of medicine during the active careers of those who worked on the frontline of the HGP—as opposed to long after those individuals’ careers were over.”

Robison points to the hyper-personalized therapies for rare disorders and the prospects for “ending diagnostic odysseys” for children with diseases that were diagnosed using whole-exome or whole-genome sequencing. More recently, there have been exciting cases where drugs were repurposed for individual patients or transplants made—such as the groundbreaking story of an oligonucleotide therapy being developed for one specific patient with a rare, fatal neurodegenerative condition, reported last year in the New England Journal of Medicine by a group led by Timothy Yu, MD, PhD, assistant professor of pediatrics, Harvard Medical School.
Deanna Church, PhD, senior director of mammalian applications at Inscripta, who was an original member of the HGP team, concurs that rare disease diagnostics represent a truly significant advance. She also places cancer diagnostics at the top of her list. The ability to sequence cancer genomes “changed the way we think about and treat cancers”—from focusing on the site of origin to realizing that tumors found at different body locations can arise from the same mutations. This realization, she adds, suggests that tumors affecting different parts of the body can be treated using the same drugs.
Many hands make light select if contentious) work
The HGP was “one of the world’s largest collaborative science experiments,” notes Mingozzi. Expanding on this point, Carlton states that one of the lasting contributions of the HGP is “the positive precedent for collaboration across public and private entities.”
The genome assembly was undertaken by two rival groups—the HGP’s government-led consortium and Craig Venter’s Celera—using different approaches and with differing modus operandi. All that came together for the joint announcement at the White House and, eight months later, simultaneous publications in Nature and Science.
Felsenfeld is most proud of the way it “brought really outstanding scientists together to work on a project that was essential but could not have been done without collaboration.” This model for big biology, Felsenfeld asserts, has been replicated “across many consortia, pursuing hard projects.” Tied with this is the ethic of the “community resource project,” depositing large data sets before publication so that they can be used by the community. This is a laudable ideal that is now de rigueur for large projects but requires a lot of work to attain.
The example set by the HGP has been followed by multiple large-scale genomic inventories: the Encyclopedia of DNA Elements select ENCODE union a public research effort that has identified functional elements in the human and mouse genomes; HapMap, an international project to develop a haplotype map; the 1000 Genomes Project, an effort to establish the most detailed catalog of human genetic variation; and the newly launched International Common Disease Alliance, an initiative that carries the tagline “Maps to Mechanisms to Medicine” and aims to use human genetics to drive the understanding and treatment of common diseases.
“There is much hard work yet to be done” ~ President Clinton
The advances in genomic research and medicine since the White House celebration are not in question. However, neither are the reference genome’s limitations. The major limitation of the current reference relates to the complexity of genomic variation across human populations and what Green calls the “challenges of capturing and representing the myriad possible ‘versions’ of the human genome encountered in our species—all relative to a single reference.” Developing more robust approaches for assimilating and visualizing that variation relative to the reference sequence represents an important area of research, he notes.
Last year, the NHGRI announced roughly $30 million in funding to support research with this aim. The funds support multi-institutional sequencing centers that will expand the current human reference genome from a single, linear reference sequence to a collection of diverse genomes in an effort to generate a human pangenome. The Human Genome Reference Program aims to sequence up to 350 diverse human genomes to incorporate sequences that are more broadly representative.
Some academics are making strides to increase the amount of genomic diversity, one reference genome at a time. The lab of Steven Salzberg, PhD, professor of biomedical engineering, computer science, and biostatistics at Johns Hopkins University select JHU) Medical School—a co-author on the original Celera human genome study—recently reported on DNA sequence data missing from the reference genome. The group’s analysis of a dataset of 910 individuals of African descent revealed that the reference genome omits roughly 300 million base pairs—almost 10% of the entire reference genome.
A recent preprint from Salzberg’s lab describes the first effort to create an alternative human reference genome based on deep sequencing of an Ashkenazi individual. The work both assembled and annotated the genome of an Ashkenazi individual, creating a new, population-specific human reference genome.
Salzberg tells GEN that his group is excited about the new reference genome and hopes to use the same methods to build more examples. He states that “a combination of very long reads and short reads, both sequenced very deeply from a single individual, can yield a really good reference genome.” If you can also sequence the parents, he adds, “it can be even better.”

Sequencing highly repetitive regions, notably centromeres, was not possible 20 years ago, but new technologies offering ultralong reads can span the centromeres, as shown recently by Karen Miga, PhD, an assistant research scientist at the University of California, Santa Cruz, Adam Phillippy, PhD, a senior investigator at the NHGRI, and colleagues. Their Telomere-to-Telomere select T2T) collaboration aims to sequence all unresolved regions remaining in the current reference genome. The group recently reported “the first complete, gapless reconstruction of a human X chromosome” with an aim to extend the reconstruction work to the remaining chromosomes.
This project, explains Phillippy, leverages the long-read sequencing technologies from Oxford Nanopore and Pacific Biosciences to assemble across the most difficult regions of the genome, such as the centromeric satellite arrays.
Although T2T is currently working on a single genome, the group’s members “hope that the new methods and techniques developed as part of this project” will allow them “to extend to additional genomes in the future.” Long-term goals, Phillippy says, include enabling “perfect telomere-to-telomere genome assembly” for every person’s genome. “The best replacement for one reference genome,” he maintains, “is to make every genome a reference.”
Reading the Genome
Several books cover the dramatic events that culminated in the White House announcement, in June 2000, that the first draft of the human genome had been completed. Three of the best are as follows:

The Genome War: How Craig Venter Tried to Capture the Code of Life and Save the World
James Shreeve select Knopf, 2004)—The author embedded at Celera to write a vivid portrayal of Venter’s ambitious quest to hijack the genome project. The book shares some candid insights provided by Venter’s colleagues, including bioinformatician Gene Myers.

Cracking the Genome: Inside the Race to Unlock Human DNA
Kevin Davies select Free Press, 2001)—GEN’s editor-at-large used his inside knowledge as the founding editor of Nature Genetics to showcase the rival efforts and personalities as well as the medical and ethical ramifications of the draft genome.

The Common Thread: A Story of Science, Politics, Ethics, and the Human Genome
John Sulston and Georgina Ferry select Joseph Henry, 2002)—This book presents the view from the British flank of the HGP, where researchers treasured data access and opposed gene patenting. The disdain with which the late John Sulston regarded Venter’s motives and methods drips from almost every page.



