February 15, 2015 (Vol. 35, No. 4)
Through Their Multifaceted Regulatory Roles, microRNAs Add Sparkle to Gene Expression
Once dismissed as so many scraps from the genomic junkyard, microRNAs are now recognized as jewels in the elaborate setting known as transcriptional regulation. In recent years, the microRNA sparkle has been glimpsed in multiple organisms, but certain facets of microRNA biology have yet to be revealed. One of the more dazzling aspects is microRNA’s multiplicity: each microRNA regulates multiple targets, and each of those targets is regulated by several, sometimes tens of distinct microRNAs.
Advances in understanding the biology of microRNAs are impacting every biomedical discipline. From basic research to clinical applications, these advances are reshaping areas including development, regenerative medicine, cancer biology, and drug delivery. Such diversity reflects the interdisciplinarity that is emerging as a defining characteristic of virtually every biomedical endeavor.
Competitive Target Inhibition
“microRNA regulation is one of the major, cross-cutting questions of a very general nature that need to be addressed as quantitatively as one can,” says Phillip A. Sharp, Ph.D., professor of biology at MIT and co-recipient of the 1993 Nobel Prize in Physiology or Medicine. Dr. Sharp’s laboratory was among the first to explore the idea that endogenous RNAs could regulate other RNA trancripts through a competitive mechanism.
This idea, called the competitive endogenous RNA (ceRNA) hypothesis, suggests that RNA molecules that share one or more microRNA binding elements can influence each other’s levels by competing for a limited pool of microRNAs. According to this model, RNA molecules targeted by the same microRNAs compete with one another and may titer away the microRNAs.
Target recognition is shaped by several variables, including target site accessibility and binding affinity. Binding affinity is shaped to a large extent by the length of sequence complementarity between mRNA targets and positions 2–7, known as the seed, within microRNAs that are incorporated into an Argonaute protein-based silencing complex. Binding affinity is highest for an 8-mer seed match, is lower for a 7-mer, and is lowest for a 6-mer.
To better understand the competition between endogenous microRNAs, Dr. Sharp and colleagues performed a quantitative analysis of microRNA networks in two cell types. In this analysis, four genome-wide microRNA measurements were used to integrate measurements of the microRNA and the target concentrations.
One of the strengths of this strategy was its use of crosslinking to identify target binding by microRNAs. “Our strategy was able to distinguish, in vivo, targets bound by an 8-mer, and showed that a 7-mer crosslinked less and a 6-mer did not, clearly indicating the existence of a gradation of the binding of microRNAs,” notes Dr. Sharp.
The findings were subsequently validated with single-cell microRNA reporter assays, which allowed the simultaneous analysis of in vivo target overexpression and the effect on microRNA activity. This approach confirmed that low microRNA-to-target ratios are sensitive to target competition and that high microRNA-to-target ratios are insensitive to target competition. It also illustrated that the differences in the susceptibility that different microRNA species show for target competition depend on the endogenous microRNA-target ratios.
Dissecting endogenous RNA competition in additional experimental settings will provide an important direction for future research. “We need to address similar questions in vivo, in cells and tissues, where the microRNA and the mRNA population have co-evolved to regulate very specific functions,” insists Dr. Sharp.
This strategy promises to unveil mechanisms that operate in those cells under physiological and pathological conditions. “For instance, that would help identify the factors that facilitate microRNA binding to a specific subset of targets in a liver as opposed to a brain cell,” explains Dr. Sharp.
Exploring the concept of competitive RNA in tumors is particularly of great clinical interest. The field of cancer genomics has revealed that tumors may have a wide diversity of mutations in oncogenes and tumor suppressor genes. These mutations often take decades to accumulate.
“If an imbalance occurred early on in the microRNA pathways, detecting those changes by genomic analyses would be difficult or impossible,” concludes Dr. Sharp. “It is important to understand the contribution of these changes and be able to rule them in or out in specific cases.”
Pluripotent States
“We found it quite remarkable that different microRNA expression profiles independently characterize different pluripotent states,” declares Thomas Preiss, Ph.D., professor of RNA biology at The Australian National University. While reprogramming somatic cells into pluripotent cells, Dr. Preiss and colleagues identified the existence of an alternate state, the F-class state.
This finding recently emerged from a study that used inducible versions of the four Yamanaka transcription factors to generate F-class and induced pluripotent stem (iPS) cells by reprogramming. In this study, Dr. Preiss and colleagues relied on next-generation sequencing to capture changes in small RNA molecules during the transitions between different cellular states.
At the time it was carried out, this study was the most comprehensive analysis of small RNA molecule expression during somatic cell reprogramming. It revealed very complex and specific patterns of microRNA expression for each of the distinct states of pluripotency.
“We added another cellular state to the reprogramming portfolio,” recalls Dr. Preiss. “This raises the concept that we may be able to characterize other pluripotent stem cell states, and that each of them may be suitable for another application in regenerative medicine.”
The possibility of experimentally manipulating microRNAs and transitioning between pluripotent states would also open new approaches to the generation of specific types of pluripotent cells. “Another, very specific use of this information,” continues Dr. Preiss, “would be to employ microRNA signatures as an identifier for one state or the other, allowing a particular profile to be used as a diagnostic.”
One of the concerns in regenerative medicine has stemmed from the fact that reprogramming uses Yamanaka factors, which can also be oncogenes. Specifically, ensuring that the end product does not contain any traces of this oncogenic potential is a key goal.
Pluripotent cells as required for regenerative medicine are functionally equivalent to embryonic stem cells (ESCs) from the inner cell mass of the early embryo. “ESCs can be taken out of their physiological context and put into specific conditions to propagate them in cell culture,” explains Dr. Preiss. ESCs that are grown in cell culture are exposed to an artificial environment that maintains their pluripotent character, but these cells are no longer exactly like the cells from the inner mass of the early embryo.
“And as we experimentally go down the path of somatic cell reprogramming,” Dr. Presiss point out, “That is an entirely synthetic process that does not really have an equivalent in nature.”
Nevertheless, exploring pluripotent states in culture is essential. These states promise to reshape regenerative medicine and many other medical fields. “And for sure we will learn more things about developmental biology during this process,” adds Dr. Preiss.
Tumor Microenvironment
“We have spent the last few years thinking about ways to better target oncogenic microRNAs or replace tumor suppressor microRNAs in cancer cells in vivo,” says Frank Slack, Ph.D., professor of pathology and director of the Institute for RNA Medicine at Harvard Medical School. One of the difficulties in targeting microRNAs to cells is that, except for the liver, and sometimes the lung, many technologies are not very efficient in reaching cells from specific organs. Several chemistries have been developed, but most of these present shortcomings.
“There has been a lot of interest over how the tumors tend to be slightly more acidic because they produce a lot of acid through glycolysis instead of using oxidative phosphorylation,” notes Dr. Slack. To take advantage of the increased acidity of the tumor microenvironment, Dr. Slack and colleagues, together with collaborators from Dr. Donald Engelman’s lab at Yale University, developed an approach to target the tumor microenvironment using a peptide with a low pH-induced transmembrane structure. This approach is denoted by the acronym pHLIP, which stands for pH (low) insertion peptide (pHLIP).
Under the acidic pH conditions found in malignant tumors, this peptide inserts into the plasma membrane and allows the antisense molecules to be transported into tumor cells, where they can target the microRNAs and specifically inhibit their function. “This opens up a whole range of new avenues for designing new drugs,” asserts Dr. Slack.
This approach provides an opportunity to deliver nucleic acids into cancer cells, where they can silence aberrantly overexpressed microRNAs. However, certain pathological conditions result from the aberrant silencing of specific microRNA molecules. “Trying to deliver microRNAs themselves as drugs, when they are lost or deleted, is a bigger challenge because RNA is very labile and more readily degradable than other chemistries that we are using,” explains Dr. Slack. “And it has to remain functional inside target cells.”
One option that Dr. Slack and colleagues are exploring is to design nanoparticles that protect the RNA in the bloodstream and deliver it to the correct cells. “But the RNAs have to escape from the nanoparticle and get into the cytoplasm to reach their intracellular target,” admits Dr. Slack. “This is still a serious challenge.”
Animal Models
“We are broadly interested in understanding the regulation and function of noncoding RNAs in mammals and how this relates to disease,” says Joshua Mendell, M.D., Ph.D., professor of molecular biology at the University of Texas Southwestern Medical Center. Most recent efforts in Dr. Mendell’s lab have focused on exploring microRNAs using in vivo animal models.
“Some of the things we thought we knew, from large bodies of work using cell lines, turned out not to be entirely correct,” observes Dr. Mendell. “[This shows] how important it is to examine the function of microRNAs in vivo, in their natural contexts.”
A recent study in Dr. Mendell’s group focused on microRNA-143/145, a cluster of two co-expressed microRNAs that has been extensively studied in human cell lines. MicroRNA-143/145 overexpression in human colorectal cancer cell lines results in very potent antitumorigenic activity.
“That has been interpreted as meaning that these microRNAs act as tumor suppressors in colon cancer,” explains Dr. Mendell. To examine the function of these microRNAs during mammalian development, Dr. Mendell and colleagues generated a mouse knockout, and one of the first surprising findings was that the mice developed normally and had a normal life span.
When wild-type mice were subjected to intestinal injury, they were able to tolerate injury well; however, the knockout mice were not able to mount the wound-healing response. “That also was surprising because the literature suggested that these microRNAs are tumor suppressors,” remarks Dr. Mendell. “But the decreased proliferation that we found in knockout mice was not consistent with a tumor suppressor role.”
Another surprising finding was that in vivo, the microRNA cluster was expressed not in the intestinal epithelial cells from which malignancies derive, but only in the supportive stromal and mesenchymal cells. “This finding contradicts the body of literature that proposed that these microRNAs are tumor suppressors intrinsic to epithelial cells, the cell type from which the tumors develop,” points out Dr. Mendell.
One explanation for the lower levels of these microRNAs in colon cancers is that the microRNA-expressing mesenchymal cells are depleted from the tumors. “It is not a decrease in the level of microRNAs [that matters],” reasons Dr. Mendell. “Instead, the tumors themselves have fewer of the cells that express these microRNAs.”
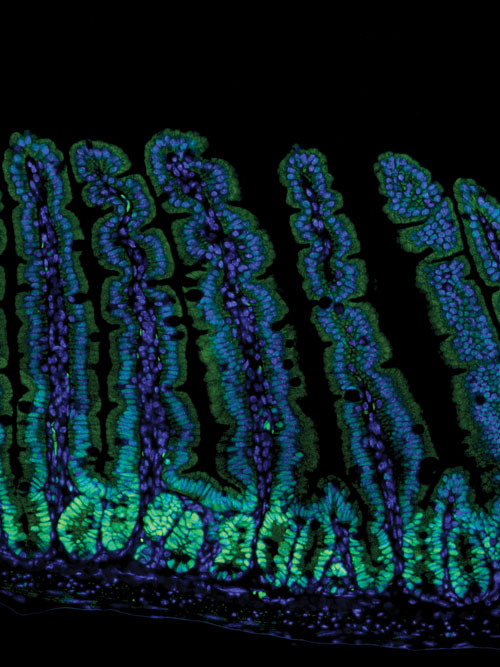
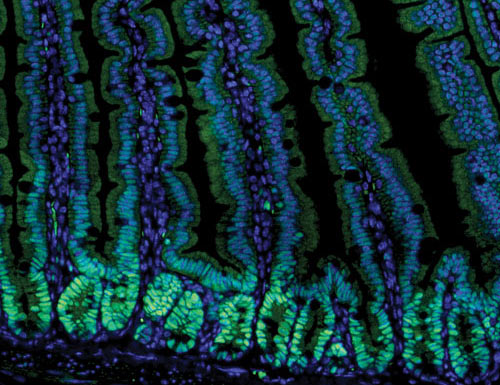
Image of the mouse intestine provided by the UT Southwestern Medical Center. Note that the tissue has been stained with an antibody that recognizes EZH2 (green) and with DAPI, which labels nuclei (blue).
Transcription Start Sites
“It is becoming increasingly apparent that we must identify the functional elements of the genome and annotate transcriptional start sites if we are to understand how the genome is built and modify it experimentally,” says Aris N. Economides, Ph.D., executive director for genome engineering technologies at Regeneron Pharmaceuticals.
Dissecting the transcriptional regulatory networks that microRNAs participate in represents one of the most challenging tasks to elucidate their involvement in health and disease. “We had the idea that knocking out the gene called Drosha, which is involved in the first step of microRNA biogenesis, would help us find the transcription start site for genes that encode microRNAs,” explains Dr. Economides.
Dr. Economides and collaborators recently developed microTSS, an algorithm that uses next-generation sequencing and can accurately predict in silico, at a single-nucleotide resolution, intergenic microRNA transcription start sites. MicroTSS scans for regions with increased RNA-Seq enrichment to identify islands of transcription located upstream of intergenic microRNAs, and it identifies the 5′ end of putative transcription start sites.
Subsequently, three independent support vector machine models—which detect histone-4 lysine-4 trimethylation, polymerase II occupancy, and DNase-Seq digestion patterns—are used to score each candidate transcription start site and complete the prediction. Dr. Economides and colleagues validated these predictions experimentally in a Drosha-null mouse model.