CRISPR technology has arguably transformed the molecular biology field more rapidly and thoroughly than any other recent advance. Using this novel gene editing technology, scientists can create new knockout animal models, target a particular genomic region for sequencing, and even edit the genomes of living organisms. It is a truly remarkable development with broad implications for the entire life sciences realm.
But the specificity of this approach has come into question. A growing body of evidence has demonstrated off-target effects caused by CRISPR edits. More recently, unsettling studies have shown that CRISPR workflows also lead to far more on-target genomic damage—that is, genomic damage made at or near the point where CRISPR editing was aimed—than researchers expected.
Until we can comprehensively identify all of the effects of genomic editing with CRISPR, we will remain limited in our ability to fully realize the potential of this technology. As CRISPR-based methods move closer to the clinic, it is critical that we establish a clear foundation for how this approach can be used in a responsible, thoughtful manner to generate the desired effects without inadvertently causing deleterious effects.
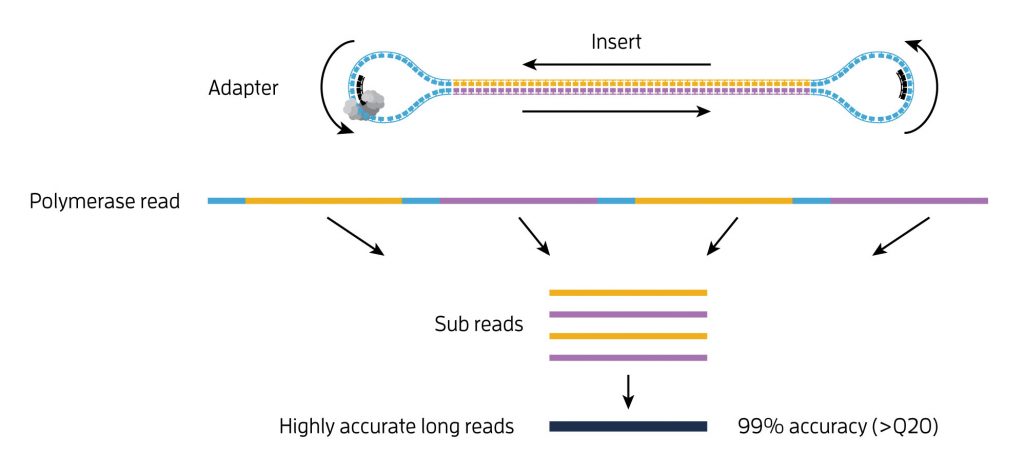
Two pioneering studies demonstrate that the extraordinarily long reads generated by Single-Molecule Real-Time (SMRT) sequencing technology can accurately detect and characterize the effects of a CRISPR gene editing experiment. This simple technique makes it feasible to verify the results of such an experiment and to look carefully for unexpected effects before moving ahead to the next step. With this validation protocol in place, scientists can now deploy CRISPR while paying close attention to the downstream effects for the most careful and responsible use of this technology.
Unexpected effects
A recent report in Nature Biotechnology from scientists at the Wellcome Sanger Institute demonstrated that unexpected on-target damage of CRISPR was far more serious and widespread than anticipated.1 In the report, which was authored by Michael Kosicki et al., the scientists indicated that they found a substantial amount of mutagenesis, including large deletions and genomic rearrangements, across the samples they analyzed: mouse hematopoietic progenitors, mouse embryonic stem cells, and a human cell line.
“Current assessments,” the report’s authors emphasized, “may have missed a substantial proportion of potential genotypes generated by on-target Cas9 cutting and repair, some of which may have potential pathogenic consequences following somatic editing of large populations of mitotically active cells.”
For this project, scientists studied a controlled environment: a 5.7-kb amplicon taken from the X-linked PigA locus found in mouse embryonic stem cells. After editing with the CRISPR-Cas9 system, they then used SMRT sequencing to analyze the entire amplicon. The long reads generated are capable of spanning and accurately resolving structural variants, repetitive regions, and other elements that can confound short-read sequencers and the assembly process required to stitch those short reads back together.
SMRT sequencing data showed that the nature of DNA repair after CRISPR editing was quite heterogeneous across the target region. Kosicki et al. noted that “the most frequent lesions in these cells were deletions extending many kilobases up- or downstream away from the exon.” The scientists also reported a de novo insertion that appeared to have come from spliced, reverse-transcribed RNA. Importantly, one of the deletions they reported removed a primer binding site that would have been used for confirmation of the CRISPR edits in a typical editing workflow.
Perhaps most telling of all was what happened when the scientists repeated the experiment. They reran the same editing, sequencing, and analysis process four times to see if the unexpected on-target changes would arise in the same way each time. The results made it clear that these effects varied widely. “Each biological replicate differed substantially, despite a large number of unique deletion events sampled, indicating that the diversity of potential deletion outcomes is vast,” the authors stated.
Measuring outcomes
Now that we have a better sense of the magnitude and frequency of unexpected changes caused by CRISPR editing techniques, it is incumbent upon us to develop robust strategies for directly evaluating the results of each and every editing experiment—especially as this approach makes its way out of the basic biology lab and toward clinical use.
A study published a few years ago in Cell Reports (and covered at the time by GEN) described the use of SMRT sequencing to quantify results of genome editing experiments.2,3 Rather than rely on measurements that require some kind of reporter system, SMRT sequencing produces long reads that allow for direct assessment of editing. According to the paper’s authors, “SMRT DNA sequencing provides a rapid, quantitative, and sensitive strategy for tracking genome editing outcomes at endogenous loci.”
Other methods typically used for this kind of post-experimental assessment, such as short-range PCR assays or short-read sequencing, are unable to provide the clear and comprehensive view of the target area that’s offered by SMRT sequencing. Generating short snippets of DNA data is not conducive to capturing large and complex edits, such as deletions, insertions, and rearrangements. To truly represent the full universe of changes caused by a CRISPR workflow, scientists need to directly report the entire genomic region of interest in a continuous sequence.
What’s next
Ultimately, CRISPR editing may become the magic bullet that allows us to cure diseases in patients. While the unexpected changes caused by this genome editing system are a setback in this technology’s progress to clinical utility, they are not necessarily a deal-breaker. Instead, this information should serve as a cautionary tale, reminding us of the need to comprehensively characterize the results of any CRISPR experiment—whether it’s at the bench or in a translational or clinical pipeline. Long-read SMRT sequencing offers a path forward. By directly measuring all DNA changes post-CRISPR, scientists can now use genome editing tools with confidence for a broad range of applications.
Jonas Korlach, PhD, is CSO of Pacific Biosciences.
References
1. Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nature Biotechnol. 36: 765–771 (2018).2. Hendel A, Kildebeck EJ, Fine EJ, et al. Quantifying Genome-Editing Outcomes at Endogenous Loci with SMRT Sequencing. Cell Reports 7(1): 293–305 (April 10, 2014).<
3. SMRT Sequencing Improves Reporting of CRISPR and Other Gene-Editing Outcomes. GEN, March 28, 2014.