October 1, 2008 (Vol. 28, No. 17)
New Technique Can Be Used to Induce Changes in a Host of Organisms
The determination of complete genome sequences for a wide variety of experimental organisms established a first step in the search for an in-depth understanding of the complex genetic functions that define living entities. Extensive genetic, biochemical, cytological, and physiological analyses are now required to correlate genome sequence with the next level of understanding of genetic function.
As such, technologies that allow researchers to routinely and efficiently edit virtually any genome—by directing mutations in a truly targeted manner—will greatly enhance the understanding of basic biology and lead to novel ways of treating human disease.
Genetic Manipulation
Early approaches to genetic manipulation of mammalian cells used ionizing radiation or chemical-induced mutagenesis to make alterations to the genome. But the random nature of these nutations represented a major limitation to studying gene function. More recent methods have enabled precisely targeted genomic editing—driven by homologous recombination (HR) between an exogenously supplied donor DNA and the specified target locus. The low efficiency of this approach, however, has necessitated sophisticated positive and negative selection strategies in order to isolate the rare targeted events.
In the early 1990s, the discovery and manipulation of zinc finger protein domains started a field of research that enabled an alternative targeted approach to genetic manipulation in higher-level eukaryotes that was over 1,000 times more efficient than the prevailing HR-based gene-targeting methodologies.
Zinc fingers are structures of about 30 amino acids held together by a zinc ion, and they determine the DNA binding specificity of the most abundant class of transcription factors found in a wide range of species. Each zinc finger—which binds to a triplet of base pairs in the target DNA—can be engineered to exhibit selectively for a specific triplet. Multiple zinc fingers can be linked to generate a larger zinc finger protein (ZFP) that recognizes a DNA sequence that is a composite of the individual finger target triplets. For instance, four zinc fingers can be assembled into a ZFP that selectively recognizes a specific 12 base pair target.
Genomic Scissors
Also of critical importance was the discovery that a zinc finger DNA-binding domain can be linked to various catalytic functional domains and so target those functions to a specified endogenous locus. Thus, by linking to a ZFP to the catalytic domain of the endonuclease FokI it is possible to create a targeted zinc finger nuclease (ZFN).
But the endonuclease subunit of FokI must dimerize in order to cleave DNA. Therefore, ZFNs are designed as a pair, to bind to adjacent sequences in the target DNA with precise sequence specificity and spacing—in essence creating a pair of very specific genomic scissors (Figure 1). Delivery to the cell of plasmids or mRNA encoding the ZFNs leads to transient expression of the nucleases and subsequent generation of a double-strand DNA break (DSB).
The power of a ZFN-induced DSB comes from the fact that a DSB triggers either of two DNA repair processes— non-homologous end joining (NHEJ) or homology-dependent repair (HDR)—that can be harnessed to evoke desired genomic alterations. The error-prone repair mechanism of NHEJ often results in localized mutations due to deletion and/or insertion of short sequences at the DSB site. The occurence of such mutations within a coding sequence typically results in disruption of functional gene expression. Thus, this pathway has proven most useful for rapidly achieving targeted gene knockout.
Alternatively, in the event that the desired outcome is insertion of a transgene into a user-specified site in the genome, a donor DNA construct can be codelivered with the ZFNs. The donor contains the transgene of interest flanked by sequences homologous to either side of the ZFN cleavage site. In the presence of this donor, the DSB is repaired via the HDR pathway, by which the transgene is effectively copied into the endogenous target site (Figure 2).
ZFN-mediated gene targeting is a powerful and versatile tool for targeted genome editing of a wide range of organisms and cells. The process of ZFN-mediated gene modification is as simple as a single transfection experiment and edits occur in as little as three days. With as much as 20% of the cells undergoing targeted genomic editing, selection markers are most often unnecessary. Once high-quality ZFNs have been constructed, isogenic stable cell lines containing the modifications can be generated in as little as a month through simple dilution cloning of ZFN-treated cell pools.
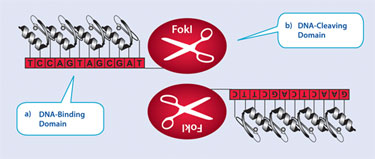
Figure 1. A zinc finger nuclease (ZFN) is composed of two functional domains
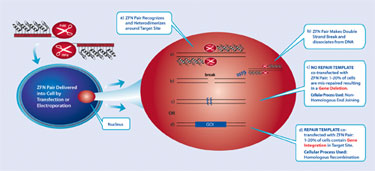
Figure 2. ZFN-mediated genome editing takes place in the nucleus when a pair of target-specific ZFNs is delivered transiently into a cell line.
Gene Knockout
Gene knockout—the complete disruption of gene function—is the most powerful tool for determining gene function or for permanently modifying the phenotype of a cell. In addition, gene knockouts can be created in multiple cell lines in order to identify and/or validate a potential drug target and provide a way to completely delete the functions of genes that are not amenable to technologies such as RNAi.
ZFNs offer a rapid single-step approach to targeted gene knockout in mammalian cells. As an example, ZFNs have been used to target the dihydrofolate reductase (DHFR) gene in a CHO cell line diploid for a functional DHFR gene. In this particular study, gene disruption at a frequency of >1% was obtained via the NHEJ pathway without the use of selection markers, and studies were done to confirm that deletion of the gene at the DNA level did indeed translate into a phenotype expected of a DHFR-null cell line.
Gene Integration
ZFN-mediated gene integration allows placement of exogenous genetic sequences precisely into user-specified sites in the genome with high-efficiency. This newfound ability will not only lead to a new generation of functional genomics studies, but it will have great impact in the drug discovery world as it will allow the creation of new cell lines containing fusion tags or reporters that can be used for more efficient cell-based screening.
In order to gain acceptance as a robust methodology, any means of targeted gene integration should occur at a frequency high enough to obviate the need for time-consuming selection of cells carrying the desired gene.
Indeed, using the ZFN methodology, targeted gene-integration frequencies of 15% and 6% have been observed, respectively, for insertion of a 12 bp tag and an 8 kb antibody expression cassette at a specific location in human cells, without using selection for desired recombinants.
In 2007, Sigma-Aldrich formed a partnership with Sangamo Biosciences to make ZFN technology available to research scientists through its CompoZr™ line of products and services.
The CompoZr custom ZFN offering, launched recently, supplies partners with zinc finger nucleases that have been validated to target and edit customer-defined genes of interest.
Future CompoZr products based on ZFN technology, available in 2009, will include ZFNs targeting popular genes, gene families, and pathways.
Conclusion
The ability to routinely edit genes in cell lines, animals, and plants has long been a goal sought after by researchers seeking an in-depth understanding of complex genetic functions and applying that knowledge to develop new pharmaceuticals, improve agricultural productivity, and even cure human disease through gene therapy.
ZFN technology promises to realize this goal by providing the tools to precisely and efficiently induce genetic changes in systems ranging from human stem cell lines to fruit flies, plants, and a host of other organisms.
Greg Davis, Ph.D., is R&D scientist, Trevor Collingwood, Ph.D., is strategic development manager, and Phil Simmons ([email protected]) is market segment manager at Sigma-Aldrich. Web: compozrzfn.com.



