October 1, 2006 (Vol. 26, No. 17)
Calorimetry Can Differentiate between Strong and Weak Binding Ligands
All biochemical reactions involve recognition, binding, and the formation of noncovalent complexes. Many small molecule drugs act by selectively binding to protein or nucleic acid targets and altering their binding characteristics. Since all binding events are accompanied by the evolution or absorption of heat (a change in enthalpy, DH), a full thermodynamic characterization of the binding reaction provides fundamental information about the molecular interactions driving the association process.
Isothermal titration calorimetry (ITC) measures the enthalpy of a reaction. If the reaction being studied is, for example, the binding of a drug candidate to a mutant protein (co-expressed with native protein due to a genetic disorder), this thermodynamic information could suggest alterations to the chemical structure of the drug, which would improve molecular interactions at the drug-protein interface, increase specificity for the mutant protein, heighten binding, or enhance efficacy of the drug.
Binding Driven by Thermodynamics
If two molecules bind, they must display greater affinity for each other than they do for the solvent. Whether or not binding occurs and how strongly and specifically, depends on the outcome of several opposing factors.
When two molecules interact noncovalently, they do so primarily through surface hydrophobic patches. Water molecules tend to be highly ordered adjacent to hydrophobic surfaces. Hence, when two hydrophobic groups interact, a large number of ordered water molecules are released into the bulk solvent. Since this increases the disorder, or entropy, of the system, this event is energetically favorable. At the same time, the two molecules lose conformational and rotational degrees of freedom. Since this decreases the entropy of the system, it is energetically unfavorable.
In addition to these entropic factors, binding is also influenced by the number of van der Waals interactions and hydrogen bonds formed or broken when the molecules interact. Clearly, binding will only occur if the sum of favorable interactions is larger than the unfavorable interactions.
The specificity of the binding reaction will be determined by various factors, including the preciseness of hydrogen bond donor and acceptor pairing, the strength of compatible electrostatic interactions, and dipole interactions.
Rational drug design requires understanding how the chemical and structural characteristics of a system affect these interactions and alter binding specificity and the integrity of the complex. Since ITC provides a full thermodynamic description of the binding reaction, the emergence of ITC as a front-line methodology in the drug development process has enhanced understanding of the correlation between thermodynamic characteristics and structural features of the interacting components.
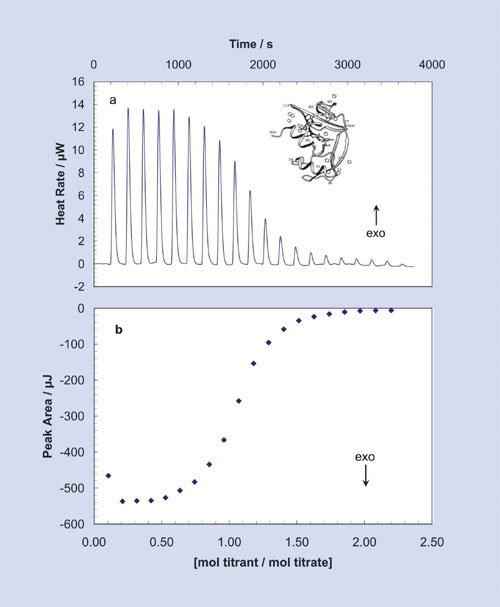
Figure 1: By using reactant concentrations appropriate for the binding reactions, raw ITC data for the titration of a small molecule inhibitor into a protein yields a sigmoidal area plot
Full Thermodynamic Description
Figure 1 shows data obtained when a total of 100 µL of 1.58 mM of the inhibitor 2´-cytidine monophosphate (2´-CMP) is titrated into 1.0 mL of 80 µM RNase A using an ultrasensitive ITC instrument specifically designed for dilute biological solutions, the 5300 Nano-ITC III from Calorimetry Sciences (www.calorimetrysciences.com). With each 5-µL incremental injection of 2´-CMP, heat is released and an equilibrium of free and bound ligand is established until, at the end of the titration, all the binding sites on the protein are occupied by the inhibitor and no further heat is produced.
The top panel of Figure 1 depicts the signal produced by the sequence of injections, and the bottom panel of Figure 1 shows the data after integration of each injection peak. The sigmoidal shape of the plot in the bottom panel of the figure, with numerous data points throughout the curved portion, facilitates determination of the midpoint of the transition and thus the stoichiometry of the binding reaction.
The association constant (Ka) and DH are calculated by iterative approximation and are performed automatically by the software supplied with the 5300 Nano-ITC III. Optimal results (pronounced curvature in the integrated peak area plot) are obtained when the concentration of the macromolecule is chosen so that:
10 < Ka[M]T <1000
where [M]T is the total concentration of macromolecule in the sample cell titrated by the ligand.
As shown in Figure 2, the relative magnitude of the equilibrium constant for a reaction with a 1:1 stoichiometry affects the shape of the titration curve, as does the concentrations of the reactants; the lower the concentrations, the larger the association constant that can be determined. A well-designed experiment, yielding good curvature in the peak area plot, typically requires macromolecule concentrations in the range of 10–100 µM, permitting Ka values in the range of 105–108 M-1 to be accurately calculated.
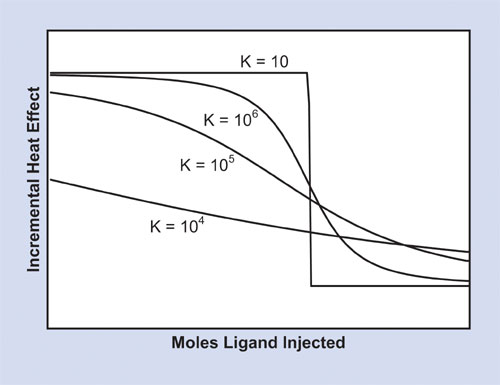
Fugure 2: Simulation of the effect of binding affinity on the shape of the titration curve for a reaction with 1:1 stoichiometry at a fixed (delta)H
Competitive Binding
The direct determination of very weak association constants can require such high concentrations of reactants that the experiment may be impractical or encounter solubility problems. Measuring very tight association constants, on the other hand, may require such low concentrations of reactants that the heat signal from the binding event cannot be measured accurately.
Very weak or very tight association constants can be conveniently determined by ITC using competitive binding. In a competition experiment, two ligands with different affinities compete for the same binding site on a macromolecule (e.g., a characterized drug and a new drug candidate competing for the same protein target binding site). Since the binding constant of the characterized drug is known, the change in its binding constant due to the presence of the new drug candidate allows the binding constant of the uncharacterized ligand to be calculated, even if its binding constant falls outside the range normally accessible by ITC.
In a typical experiment to measure the Ka of a weakly binding ligand, for example, sufficient weak ligand is added to the target macromolecule to occupy approximately 50% of the binding sites. Using ITC, the binding sites are then incrementally filled by the characterized, stronger-binding ligand, displacing the weaker ligand until the protein is saturated. The extent to which the second ligand displaces the first will be dependant on the relative affinities and concentrations of the competing compounds.
A similar approach can be used to measure the Ka of very strongly-binding ligands. An application of the competitive binding approach for determining the Ka of a weakly binding inhibitor is presented in Figure 3. RNase A binds both 2´-CMP and 5´-CMP in the same binding pocket, but shifting the hydroxyl group from the 2´ to the 5´ position significantly decreases the binding affinity of the ligand. The single binding site of RNase A is so weakly inhibited by 5´-CMP that there is little curvature in the titration plot, so the affinity of 5´-CMP can only be roughly estimated from a direct titration experiment without resorting to high reactant concentrations (red dataset, Figure 3).
However, if sufficient 5´-CMP is added to RNase A to occupy about half the binding sites (in this example, 1.0 mL of 70-µM RNase A prebound with 0.32-µM 5´-CMP), and then 2´-CMP is titrated in (100 µL total of 1.3-mM 2´-CMP, titrated in 5-µL increments), significant heats of binding are obtained (blue dataset, top panel). This provides a graph of integrated heats (bottom panel) that can be accurately fit to provide the stoichiometry (n = 1), binding constant (3.1 x 103), and enthalpy (-47 kJ/mol) of the 5´-CMP binding reaction.
ITC is rapidly becoming the method of choice for characterizing binding reactions. The approach is completely general: small molecules binding to proteins, DNA, RNA, and polysaccharides can all be studied in an analogous manner, as can the binding of one macromolecule to another (by placing the second macromolecule rather than a small molecule ligand in the syringe).
The increasing prominence of ITC as a fundamental tool in biomedical research is due to the accurate assessment of binding interactions that can be obtained in several 1–2 hour automated experiments using just nanomoles of material and without the need to develop new assay protocols for each biomolecule or ligand. In addition, ITC’s direct nature and high precision allows users to validate the results of less rigorous, high-throughput assay protocols.
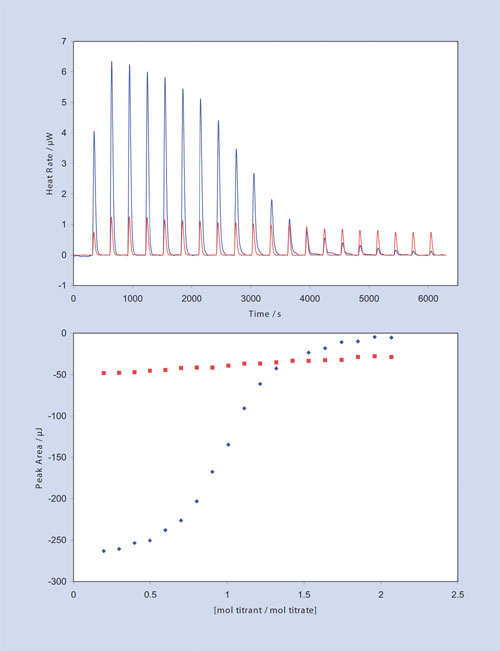
Figure 3: Competition experiments (blue) allow acurate dterminatin of the binding characteristics of a weakly bining ligand that cannot be quantified in the absence of a competitor (red)
Christin Choma is senior biophysicist, Calorimetry Sciences. Web: www.calorimetrysciences.com. Phone: (801) 763-1500. E-mail: christin.choma@ calscorp.com.



