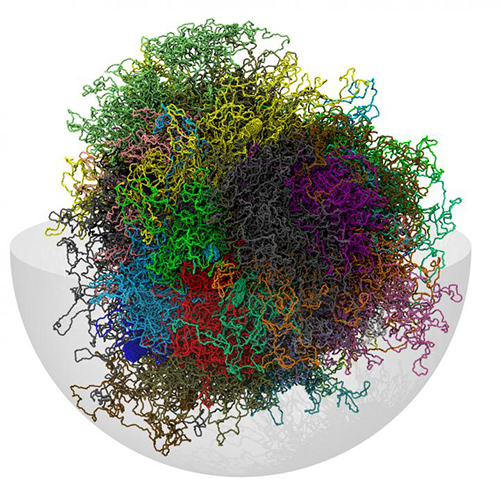
![Simulation of 3D chromosomes structure. Chromosomal spatial arrangements were determined by using Hi-C-constrained physical models. [SISSA]](https://genengnews.com/wp-content/uploads/2018/08/Oct27_2016_SISSA_3DChromosomesStructure8029163462-1.jpg)
Simulation of 3D chromosomes structure. Chromosomal spatial arrangements were determined by using Hi-C-constrained physical models. [SISSA]
A shortcut has been found that may hasten the arrival of a genomics that is not just sequence-smart, but also shape-smart. The shortcut is possible because of proximity pairs, known chromosome–chromosome contact propensities, which may be catalogued by means of the Hi-C chemical–physical technique. By accounting for proximity pairs, researchers based at the International School of Advanced Studies (SISSA) had a means of constraining the various ways chromosomes could pack together. Ultimately, these researchers, led by Cristian Micheletti, created a 3D genomic map that captured structural features that had been observed in vivo.
The SISSA researchers suggest that their technique may help advance efforts to understand the organizational principles regulating the human genome. They described their technique in a paper (“Hi-C-Constrained Physical Models of Human Chromosomes Recover Functionally-Related Properties of Genome Organization”) that appeared October 27 in the journal Scientific Reports.
“First, we use advanced statistical tools to single out local and non-local cis-chromosome contacts that are significantly enriched with respect to the reference background of Hi-C data. Second, we employ steered molecular dynamics simulations to drive the formation of these constitutive contacts in a physical model of the human diploid genome, where chromosomes are coarse-grained at the 30 nm level. The viability of this general strategy is here explored for two distinct human cell lines: lung fibroblasts (IMR90) and embryonic stem cells (hESC).”
Essentially, the SISSA researchers used experimental data on proximity pairs to arrive at a precise description of the shape of the incredibly complicated tangle of DNA that is the genome.
“Imagine having to create a map of a city,” explained Micheletti, “based only on information like 'the post office is opposite the station,' 'the chemist is close to the gym,' 'the fruit and vegetable market is near the football field,' and so on. If you have only a small number of such statements to go by, your map will be approximate and in some cases indeterminate. But if you have hundreds, thousands or even more, then your map will become increasingly precise and accurate. This is the logic we followed.”
Proximity pairs refers to information on the closeness of two points on the map. In the case of nuclear DNA, this information was the Hi-C technique, which was developed by North American research groups in 2010. With Hi-C, bits of genome located close to each another in the nucleus are tied together and then identified by their sequence. By collecting large numbers of these proximity pairs, scientists discovered which points of the chromosomes lie close to each other in the nucleus. While this is today the most powerful technique for investigating DNA organization in the nucleus, it is still inadequate for inferring its overall shape.
“For this reason, we thought we would try to go 'further,'” commented Micheletti. The SISSA researchers created models that remain minimally entangled and acquire several functional features that are observed in vivo and that were never used as input for the model.
“We find, for instance, that gene-rich, active regions are drawn towards the nuclear center, while gene poor and lamina associated domains are pushed to the periphery. These and other properties persist upon adding local contact constraints, suggesting their compatibility with non-local constraints for the genome organization.”
The authors concluded, “We expect that the general approach followed in this study could be profitably extended and transferred to other systems, so to incorporate additional knowledge-based information and to capture more detailed aspects of the spatial organizational principles of eukaryotic nuclei. These will be even more relevant when missing genomic repeat structures eventually become incorporated into analysis. Furthermore, we in particular envisage that extending considerations from cis– to trans-chromosome contacts may be important towards pinning down more precisely the relative positioning of chromosomes, and also their absolute position in the nuclear environment.”