December 1, 2008 (Vol. 28, No. 21)
Novel Technique Allows Epigenetic Scanning Across Multiple CpG Sites
Methylation-sensitive (MS) Hi-Res Melting® is a sensitive detection method that requires no post-PCR sample manipulation. Analysis is fast and simple and achieved by comparing melting temperature (Tm) and melting curve shape. This technique is sensitive enough to show a single methylation event within a group of multiple CpGs, and multiple CpGs can be simultaneously analyzed. In addition, high discrimination is possible because of the sensitivity of Hi-Res Melting.
We amplified a 152-bp genomic fragment of a proposed regulatory element to miRNA-195 containing five CpGs distributed throughout the amplicon. We were able to differentiate overall methylation from 10 to 100% as well as see small melting differences between 39 and 40%.
Individual CpG sites appear to affect melting curve shape as does the average methylation. We also investigated ways to overcome common challenges. First, there can be variation between replicate conversions of the same sample. Commercial kits appear to minimize this problem. Second, it is commonly believed that bisulfite-treated DNA rapidly degrades, necessitating testing within hours or days of treatment. Our results demonstrated sample stability for three months at 4°C in TE buffer. Third, small sample-to-sample temperature, volume, and PCR buffer differences can confound the interpretation of results. The use of internal melting temperature calibrators during amplicon denaturation mitigated these limitations and provided for more consistent sample interpretation.
High-resolution instrumentation and advanced dyes such as LCGreen® Plus can be used to generate a highly detailed picture of methylation within a given PCR fragment. The post-PCR technique of Hi-Res Melting can be performed on bisulfite treated genomic template. This converts 5-methylcytosine to uracil, which base pairs with adenosine during PCR and, ultimately, converts to T.
Percentage differences in GC are thus transferred into the amplicon altering Tm. Hi-Res Melting is useful for detecting sequence heterogeneity. This is because heteroduplexes created from sequence heterogeneity broaden the shape of the curves. The CpG position relative to the position within the amplicon can also affect shape. Both Tm as well as curve shape can be used in combination for epigenetic studies. Methylation is inherently heterogeneous, and Idaho Technology’s LightScanner instrumentation and software with calibration are ideally suited to discriminate fine differences in sequence heterogeneity.
Materials and Methods
Standard DNA extraction was performed using commonly available kits. Bisulfite conversion on sample DNA was performed using the EpiTect™ kit (Qiagen). For hypermethylated controls, CpGenome™ Universal Methylated DNA (Millipore) was used. We also obtained DNA from Raji cells using standard kit extraction. PCR was performed using High Sensitivity Mastermix (Idaho Technology) containing LCGreen Plus, oligonucleotide calibrators, dNTPs, hot-start enzyme, and buffers.
Oligonucleotide primers were synthesized and included in the reactions at 0.25 µM each. PCR was performed on iCyclers (Bio-Rad) with an initial two minute denaturation at 95°C, followed by 45 cycles of 30 second holds at 94°C and 66°C.
Heteroduplex generation was performed on the iCycler by programming a final denature at 95°C/30 second and anneal at 28°C/30 second. Hi-Res Melting was performed on the LightScanner between 50 and 95°C. Calibration was performed, and data was analyzed in the small amplicon module using Call-IT® algorithms in the LightScanner analysis software. After melting, products were column-purified and sequenced using standard techniques.
Results
Average methylation was calculated by estimating the relative C:T peak heights in sequencing electropherograms. Total percentages for the forward and reverse sequencing reactions from each sample were summed and divided by five (the number of CpG sites within the amplicon) resulting in the average methylation. Colors indicate sample identities and are consistent throughout this article (Figure 1).
Four findings were made. First, amplicon Tm increases with average methylation. Second, the use of internal oligonucleotide-based calibration improves the Tm correlation with average methylation. This enables more accurate sample measurement and comparison (Figure 2). Third, derivative melting curve shape broadens with increasing heteroduplex content, which is a function of original 5-methylcytosine heterogeneity. This can be seen in Figure 3. Finally, repeatability between bisulfite conversions on the same sample is good using Qiagen EpiTect kits. In addition, three-month stability of bisulfite converted DNA in TE;1X at 4°C is excellent. Finally, real-time PCR results showed no relationship between crossing point and sample storage time, indicating good template stability (data not shown).
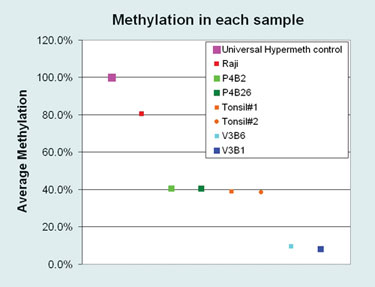
Figure 1. Distribution of the average methylation found across samples used in this study.

Figure 2. Tm is explained by methylation.
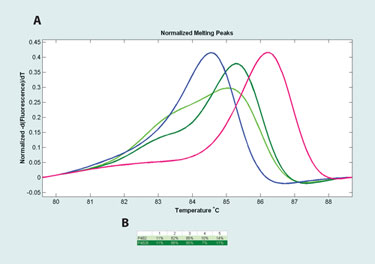
Figure 3. Uncalibrated derivative melting curves of several tumor samples and hypermethylated control.
Conclusions
Hi-Res Melting in the presence of internal oligonucleotide calibrators results in significant qualitative and quantitative information from Tm and shape. We obtained good sample repeatability between bisulfite-treated template DNA. In addition, sample stability of bisulfite-converted template at 5–15 nanograms/µL appears to be adequate beyond three months at 4°C. Using this technique, it is feasible to obtain rapid, cost-effective epigenetic scanning across multiple CpG sites.
Cameron N. Gundry (cameron_gundry@ idahotech.com) is research scientist, Mikeal Wall is customer support specialist, Jason T. McKinney is manager of the life science division, Colleen Hough, Ph.D.,
is research scientist, Lindsey Cutler is research associate, and David H-F. Teng, Ph.D., is director of R&D chemistry at Idaho Technology. Web: www.idahotech.com. John D. Phillips, Ph.D., is research associate professor of pathology and hematology and Margaret K. Yu, M.D., is assistant professor in the department of hematology at the University of Utah.



