May 15, 2012 (Vol. 32, No. 10)
“ELN” Facilitates Construction, Management, and Dissemination of Key Pathways
The fate of new chemical entities in the pharmaceutical discovery and development pipeline hinges on biotransformation pathways and the outcome of metabolite safety profiles. How best to secure and share knowledge gained from metabolite projects is a question faced by all organizations considering the disconnected nature of data acquisition and project efforts across multiple teams.
Relating biotransformation pathways to analytical and chemical information, typically generated by different research groups, often in geographically separated locations, has previously been a major challenge. Lengthy and research-intensive metabolite projects increase the risk of losing valuable information as people move from project to project or leave the organization, further hindering drug development.
A novel approach to knowledge management of metabolism studies utilizes a platform that enables organizations to link, store, and access analytical and chemical data in ways not previously possible.
ACD/Labs’ knowledge management solution for metabolism acts as a “biotransformation ELN” that combines the ability to create interactive biotransformation maps with advanced interpretation of analytical data including NMR, MS, and chromatography, and the ability to store the chemical context behind the interpretations.
In addition, measurements or predictions of chemical information such as physichochemical, ADME, and toxicity properties can be incorporated. Perhaps most importantly, the biotransformation ELN provides a means to increase the depth and richness of discovery reports and IND applications by unifying information and making it readily accessible, leading to greater insight into metabolites and metabolism (Figure 1).
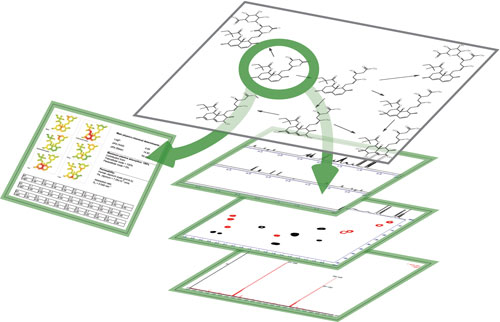
Figure 1. Analytical and chemical knowledge management helps use legacy knowledge to provide insight into current work.
Building Interactive Maps
The determination of metabolite structures and creation of biotransformation schema usually begins with the acquisition of analytical data, particularly from LC/MS instruments. Processing the data using advanced extraction and interpretation algorithms, such as ACD/IntelliXtract, can facilitate finding and characterizing metabolites.
Proposed structures, pathways, analytical data, and spectral interpretations are then readily retained and can be effectively managed, visualized, and searched as database records. Individual records can accommodate single structures or complex pathways with multiple steps of metabolism sharing common structures.
Additional data from supporting analytical techniques as well as chemical and biological data can also be associated with chemical structure, accessible to review with a simple mouse click. The stored spectra are retained with interpretations—not just as archived data—allowing the features of the spectra to be readily investigated at any time (Figure 2).
Chemical structures are paramount in metabolism studies; however, in practice, partially elucidated structures are common. The biotransformation ELN provides a means of recording discrete and indeterminate structures, relating associated data to both. For example, displayed in Figure 3 is a Markush representation of the possible desmethyl metabolites of dextromethorphan. When the site of modification is established at either the nitrogen or oxygen, one can edit the structure and continue associating analytical and supporting data as required.
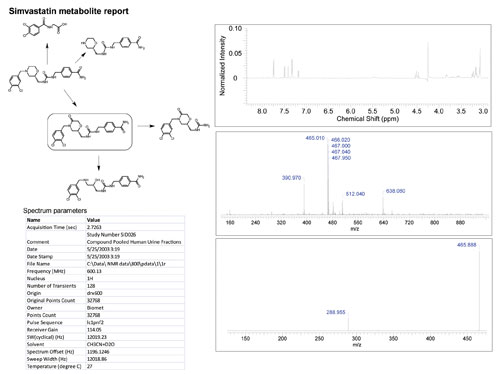
Figure 2. One view of the biotransformation ELN interface displaying an example biotransformation schema for a compound previously reported in the literature with an associated proton NMR and mass spectrum.
Legacy Knowledge
Legacy knowledge can help speed up interpretation, avoiding duplication and misleading clues to elucidation, by applying information previously gathered to current and future projects. The biotransformation ELN permits queries that reveal if a particular metabolite has been encountered in the past. Keeping structures, analytical data, supporting data, and the relationships between them means positive search results make an entire body of work immediately available for re-examination.
Structures and reactions in the biotransformation ELN are searchable in a variety of ways. For instance, search a particular structure against the database to find if it has been proposed in previous experiments or projects. Alternatively, a partial structure can be selected and a substructure search performed, resulting in a visual display of all relevant structures in the schema.
Structure searches may be confined to look only for parents/reactants or metabolites/products, and may be preceded or followed by searches of other data fields. Thus, one can readily focus on information for a particular species or biological matrix, and look to support tentative structure assignments by searching for similar reactions among legacy information.
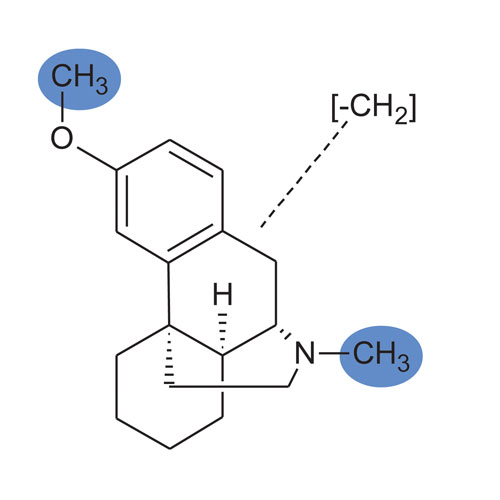
Figure 3. A Markush structure can be used to keep track of a chemical entity even if a specific site of metabolism is undetermined.
Reporting and Sharing Metabolism Knowledge
The vast amount of data collected around biotransformation schema can be extremely challenging and time-consuming to distill into a useful format such as a discovery report or IND application. The biotransformation ELN assists this process first by providing a logical and organized way of storing, viewing, and interpreting data, and then by creating professionally customized electronic reports with just a few mouse clicks.
The report can include all structural information along with formatted analytical data, including spectra and chromatograms if desired, and the scientist’s notes to provide clear communication.
Predicting Metabolism and Identifying Safety Concerns
Identifying safety concerns and detrimental physical properties for drug candidates and potential metabolites helps avoid compounds with liabilities. The biotransformation ELN also allows measured or predicted physical properties and assay results to be stored with structures in the biotransformation knowledgebase.
Predicted risk associated with metabolite vulnerabilities can be used to help redesign and select more promising candidates for further and more rigorous testing. Hepatic metabolism by cytochrome P450 enzymes is the major clearance route for xenobiotics.
Using in silico tools such as ACD/Labs cytochrome P450 predictive modules, one can identify whether compounds will be substrates and/or inhibitors of the five major drug metabolizing enzymes—CYP3A4, CYP2D6, CYP2C9, CYP2C19, and CYP1A2. Furthermore, the software predicts the most likely sites of metabolism in human liver microsomes.
These predictors also offer the ability to incorporate experimental data, which extends chemical space coverage and serves to improve prediction accuracy as results accrue. Since biotransformations can lead to either an increase or decrease in toxicity, accessory data about metabolites is an interesting and often vital part of complete compound assessment.
The End Game
Overall, ACD/Labs biotransformation ELN enables the finding and reporting of new knowledge more quickly and with greater depth; increasing the efficiency of metabolism studies. Unique ways of linking analytical, chemical, and structure information leads to improved project management, increases the confidence in structural elucidations, enables the incorporation of related predicted safety data, and facilitates the preparation of discovery reports and IND applications.
The biotransformation ELN supports integration with existing knowledge management systems and ERPs, contributing to an organization’s retention of metabolism information such that legacy knowledge can be leveraged to identify metabolites faster and to make better decisions around the safety and fate of compounds.
Graham A. McGibbon, Ph.D. ([email protected]), is product manager, mass spectrometry, at ACD/Labs.



