October 1, 2009 (Vol. 29, No. 17)
Flow Cytometer Aims to Improve Upon Traditional Methodologies
Technology that gives researchers the ability to quickly and precisely determine the number of cells with a given phenotype in cell cultures or fresh tissue samples is of great interest in diverse fields, including clinical research and diagnostics, drug discovery, and molecular biology. In this article, we present information on the function and use of the Accuri® C6 Flow Cytometer® System from Accuri Cytometers, which can quickly and reproducibly provide single-platform cell counts on fluorescently labeled subsets in mixed cell populations.
For years the combination of a light microscope and hemacytometer have been the tools of choice for performing cell counts in life science research laboratories. This method is slow and prone to error. When hemacytometer counts and flow-cytometric phenotypic data are combined to determine cell subset numbers, errors are multiplied. Digital flow cytometers, utilizing laminar-flow fluidics, allow fast, phenotypic data collection (at rates up to 10,000 events per second) on a wide range of cell types (submicron-sized bacteria through large mammalian cell lines), but still require the addition of counting beads to each sample to calculate cell-subset concentration.
On the other hand, flow cytometers with syringe-driven fluidics can deliver absolute count measurements without the addition of counting beads to samples, but are often limited by lower data acquisition rates (
The Accuri C6 Flow Cytometer System is a small flow cytometer that has a unique peristaltic-pump-driven, laminar-flow fluidics system (Figure 1). The C6 combines the advantages of hydrodynamically focused cell sampling (high data-acquisition rates, good light scatter and fluorescence resolution) with the ability to report absolute counts for any identified population in a sample.
Two applications of direct cell-concentration determination with the C6 are presented here: viability assessment of cultured cell lines and platelet counts for whole, human peripheral blood samples. Both methods make use of the fluorescence and light-scatter measurements possible with a flow cytometer to identify sub-populations of interest. Cell-counting data collected by at least one other traditional method (counting beads or hemacytometer) is included for comparison.

Figure 1. The Accuri C6 Flow Cytometer System
Instrumentation
The C6 was validated, before data collection, for accurate volume measurement using Accuri Volume Validation Beads, fluorescent beads prediluted at a known concentration.
CFlow® Plus software was used for cell-count data collection. A fluidics setting of medium (flow rate=35 µL/min, core size= 16 µm) was used during sample collection.
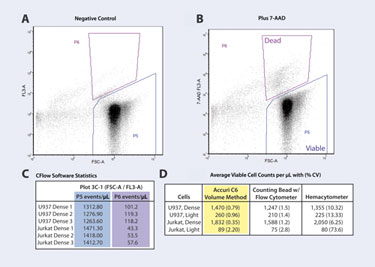
Figure 2. Viable cell-count determinations with the C6: (A) Setting viable-cell gate (P5). (B) Setting dead-cell gate (P6). (C) CFlow Software Statistics Table. (D) Comparison of mean viable cell count.
Viability Assessment
Method and Results Two common cell lines, U937 and Jurkat (available through ATCC), were used. Initial cultures for each line were seeded at both a high and low density, so that the final cell concentration at analysis would be dense or light, respectively. After one week of culture, cells were resuspended and 1 mL transferred directly from each tissue culture flask into a 1.5 mL tube. 7-Amino-actinomycin D (7-AAD, Cayman Chemical), a fluorescent dye which is excluded by viable cells, was used as a marker of cells with compromised outer membranes.
Five (5.0) µL of a 1 mg/mL stock 7-AAD solution, along with 50 µL of AccuCount Fluorescent Particles (Spherotech; ACFP-50-5) were added to each sample tube and mixed thoroughly. Tubes were kept at room temperature (RT) in the dark, and sampled between 5 and 30 minutes after addition of 7-AAD, with gentle mixing immediately prior to analysis.
A hemacytometer was used to perform the microscopic cell counts. Appropriate dilutions of cell samples were made into phosphate-buffered saline containing Trypan Blue. Counts were performed in triplicate. At least 100 nonblue cells were counted for each replicate.
With the C6, only a single 2-D density plot of forward light scatter (FSC-A) versus 7-AAD fluorescence (7-AAD FL3-A) was required for data analysis. A negative control sample for each cell type (no added 7-AAD) was used to define the viable-cell gate (Figure 2A, gate P5). This gate included events with high FSC-A, and defined the FL3-A background fluorescence. The dead-cell gate (Figure 2B, gate P6) was set using a cell sample containing 7-AAD. This gate only needs to be set once for each cell type within a given experiment.
Using the Statistics Tab in CFlow, the events/µL for these two gates were displayed for triplicate samples (Figure 2C). This data was copied and pasted into a spreadsheet program to determine the standard deviation (SD) and coefficient of variation (CV) for triplicate measurements (Figure 2D).
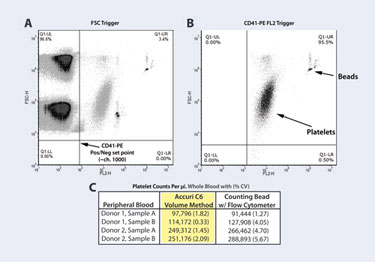
Figure 3. Platelet-count determination with the C6
Small Particle Measurements
Method and Results Aliquots of whole blood (1 or 2 µL), collected in sodium citrate tubes, were diluted 1:10 into HEPES-buffered saline with 1% formaldehyde. Twenty (20) µL aliquots of diluted blood were incubated in 1.5 mL tubes at RT, 20 minutes, with 20 µL of CD41-PE antibody (DAKO clone 5B12). Samples were then diluted with 1 mL HEPES-buffered saline with 1% formaldehyde. Five (5) µL of RFP-50-5 beads (Spherotech) were added to allow comparison of two counting methods. Samples were well mixed and read, without washing, on the C6.
For this assay, data collection was triggered by the positive fluorescence signal of CD41-PE labeled platelets (read in FL2-H), in order to improve discrimination of platelets from debris and to increase counting accuracy. The appropriate FL2-H threshold channel was determined by first triggering on FSC-H, determining where CD41-PE+ events fell relative to the negatives, and setting this value (FL2-H channel=1,000) as the primary threshold in CFlow Plus (Figure 3A). All subsequent samples were collected with this threshold in place (Figure 3B). Platelet counts per µL of sample were copied from the CFlow Statistics Tab into a spreadsheet program, and dilution factors were applied to determine the platelets per µL of original whole-blood sample (Figure 3C).
Conclusion
No statistically significant differences were found between the volumetric C6 cell concentration measurements and those obtained with standard methods (hemacytometer and counting beads) for two multiparametric analyses: viable-cell concentration for cell lines and platelet numbers per µL of original whole-blood sample.
However, the precision of the cell-count data obtained by C6 volume measurement was significantly better than that obtained by hemacytometer (pFigure 4). The average CV for triplicate cell counts with the C6 was 1.2%. The average CV for counts using counting beads was twice as high (2.8%) and that for hemacytometer counts was 20 times higher (25%).
The sources of error inherent in hemacytometer counts are well known and it is not surprising that variation between replicate sample counts would be high. The use of counting beads in flow-cytometry experiments has largely replaced combining hemacytometer counts with flow-cytometry data as a way to determine cell concentration of phenotypically diverse populations. But even this approach, as demonstrated here, is likely impacted by sources of error such as pipetting technique and calibration, inherent variability in bead-stock concentrations, and the subjective setting of a bead gate in the flow cytometer data file.
An added benefit of obtaining event counts per µL directly from the C6 data is the comparative ease of use. No complicated back calculations to determine the volume sampled based on bead number collected are required, nor are subjective decisions about how to gate on the singlet bead population.
The unique peristaltic pump fluidics system of the C6 allows direct measurement of the volume pulled from each sample tube, yet also provides standard laminar flow, which is crucial to accurate flow-cytometric fluorescence and light-scatter measurements.
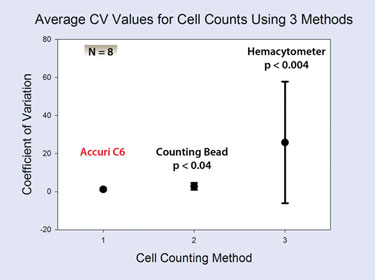
Figure 4. The average coefficient of variation +/- 1 SD for replicate cell counts using three different counting methods on the same samples
Clare Rogers ([email protected]) is applications scientist at Accuri Cytometers. Web: www.accuricytometers.com. Erin Gatza, Ph.D., is senior scientist at Lycera.



