October 15, 2010 (Vol. 30, No. 18)
Nonlinear Path from Discovery to Validation Remains a Problem
The discovery of objective biomarkers for diagnostic and therapeutic purposes is the subject of a growing body of research. Particularly important are those biomarkers that predict disease initiation/recurrence or objectively measure the response to treatment in patients without outward physiological symptoms.
Translation of biomarkers along the biomarker pipeline into the clinical practice is not a linear process, however. Despite many candidate biomarkers identified in the scientific literature, only a few have been validated. This is largely attributed to the high costs of clinical validation studies.
The National Cancer Institute (NCI) has taken a lead role in supporting biomarker translation. In 2006 the NCI launched the Clinical Proteomic Technologies for Cancer (CPTC) initiative. This initiative introduced an intermediate step in the biomarker pipeline called biomarker verification.
According to the agency, “verification, the bridge between discovery and qualification, is the process of credentialing prioritized biomarker candidates using analytically robust, reproducible, and quantitative assays on a statistically powered number of samples with clinical relevance. Credentialed proteins successfully passing this stage are considered verified biomarkers, which are of high value for translating into large-scale clinical qualification studies.”
The presence of such a bridge can rapidly triage the candidate biomarkers prior to investing large sums of money and time on development of a commercially suitable assay. Such a path could also enable verification of new technologies.
While ELISA is still a gold standard for diagnostics, the limitations of this technology become more and more apparent as scientific attention is drawn to the biomarkers present in low concentrations, represented by complex signatures, or arise via isomeric changes. This article describes several cutting-edge technologies bridging the biomarker pipeline from discovery to verification.
Disease-Relevant Protein Isomers
Protein variants, such as conformational isomers, post-translational modifications, point mutations, and degradation products, represent a rich resource of diagnostic information that has remained unexploited. These isoforms are normally present at low concentrations, and often cannot be differentiated by antibodies alone.
At GTCbio’s Biomarker Discovery and Development Conference, which will be held next week in San Francisco, scientists will discuss a range of approaches for working with protein biomarkers. For example, Intrinsic Bioprobes is developing mass spectrometric immunoassays (MSIA™) for clinical diagnostics.
“Our goal is to translate the MSIA technology into routine clinical applications,” says Urban Kiernan, Ph.D., director of biomarker discovery. “MSIA occupies a unique niche in diagnostics—quantification of disease-relevant protein isomers. Even if antibodies are unable to separate truncated forms or point mutations of the same protein, the combination of affinity-capture with MS provides a powerful diagnostic tool.”
The Massay® platform supports the fully automated process that begins with the capture of the target analytes from crude biological fluids by specific antibodies. The capture antibodies are immobilized on a proprietary solid support fitted within functional pipette tips. Each antibody is empirically selected for the desired target.
During the sample incubation process, biological fluid is recirculated through the affinity tip for enhanced enrichment of the desired target. The captured analyte is then eluted and directly deposited onto a mass-spec target for MALDI-TOF MS analysis. Integrated robotics enables high-throughput processing of 96 samples at a time.
“Research behind clinically relevant protein isoforms had been lagging until we pioneered a robust way to quantify protein variants,” continues Dr. Kiernan. “Just by screening the normal population, we were able to identify a substantial number of new isoforms. This opens the door to a completely new branch of clinical proteomics.”
One of the company’s early successes came from the analysis of brain natriuretic peptide (BNP), a well-known marker of congestive heart failure. The BNP assay has become one of the most important blood tests in cardiology. And yet, the available antibody-based test is unable to distinguish between circulating BNP forms, some of which are not clinically relevant but add to the total BNP measured.
Massay technology not only detects such low-abundance analytes but enables the unequivocal quantitation of active and inactive forms, ultimately driving the choice of therapeutic intervention. The company has also recently discovered and qualified two isoform biomarkers for type 2 diabetes that have been licensed to Ortho-Clinical Diagnostic.
“This approach makes sense,” comments Lawrence Oliver, Ph.D., professor in the department of laboratory medicine and pathology at the Mayo Clinic. “A number of clinical biomarkers are known to be present in different forms. For example, IL-6 is present in 17 different isomeric forms. This complicates our ability to consolidate the results of various clinical trials measuring the same analyte.”
Dr. Oliver is proposing a study that would re-evaluate the clinical validity of a well-established biomarker, an indicator of increased risk of coronary disease and stroke. Lipoprotein-associated phospholipase A2 (Lp-PLA2) generates pro-inflammatory products as a result of its natural enzyme action and is both a biomarker and a therapeutic target studied in multiple trials of vascular disease prevention.
Most recently, The Lancet published the retrospective analysis of 32 clinical studies with over 79,000 participants. The meta-analysis of this data demonstrated association of Lp-PLA2 activity and mass with risk for coronary disease. However, Dr. Oliver demonstrated that this analyte is not only unstable at room temperature, but also at –20ºC.
In his studies, the values for Lp-PLA2 stored at –20ºC steadily rose by as much as 40%. Such drift was not observed at –70ºC. Freeze-thaw cycles increased values by as much as 18%. Mayo Clinic collaborated with diaDexus, the manufacturer of PLAC® test for Lp-PLA2, on validation of an automated test.
“diaDexus had to overcome significant issues with the reagents,” continues Dr. Oliver. “Their newest test meets analytical validation criteria. But the absolute values of Lp-PLA2 detected by the new version of the test are only at 50% of what was published in the previous 32 studies.”
The contrasting values may be due to the drift under low temperature storage conditions, or due to differential stability of the enzyme in distinct states of the disease, or because the new test differentiates between bound and unbound forms of Lp-PLA2.
Regardless of the cause, the new test may not be measuring the risk component to the same extent as the old one. This means that the prospective data cannot be correlated with any previously collected clinical data. “This very promising biomarker can no longer be supported by the previous body of data,” adds Dr. Oliver. “Whenever a new biomarker is discovered and put to clinical use, the manufacturer has to ensure that with every new iteration of the test, we can continue to trace the results back to the clinical trials used to validate its interpretation.”
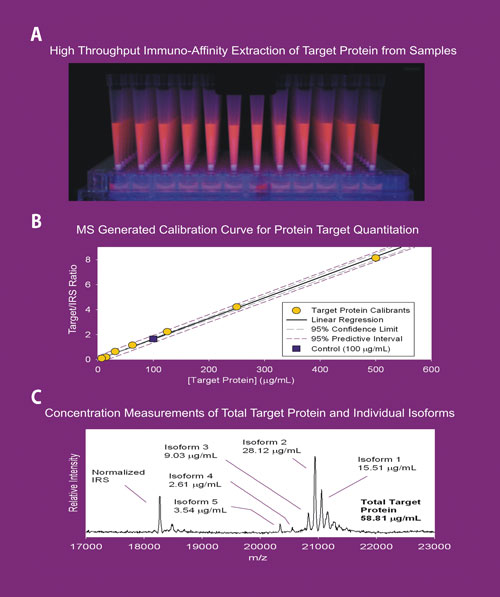
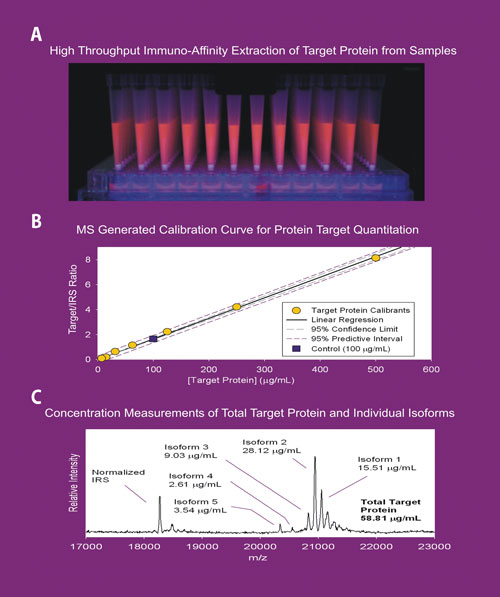
(A) High-throughput application of MSIA™ technology using the MASSAY™ platform. (B) A calibration curve generated for quantitative MSIA application. (C) MSIA analyses for measuring target protein concentration. According to Intrinsic Bioprobes, this approach provides the ability to determine the individual concentrations of target protein variants that constitute the total protein value.
Epigenetic Biomarkers
Just about any part of the biological continuum from DNA to metabolites is being mined for novel biomarkers. Among them, DNA-methylation patterns are gaining more and more attention because of their broad implication in human disease. Epigenetic signals present a complex language that we are yet to understand.
Since these signals are directly linked to DNA, they are believed to be causative rather than consequential of many downstream processes. In cancer studies, many lines of evidence point to the epigenetic contribution to cellular abnormalities. However, finding and understanding the patterns of epigenetic language is not trivial.
Orion Genomics has developed several tools to efficiently and comprehensively measure epigenetic signals for the entire genome. MethylScope® reportedly enables genome-wide DNA-methylation profiling. Simultaneous study of the epigenetic state of every human gene as well as hundreds of thousands of other loci enables detection of candidate biomarkers that otherwise would remain undiscovered.
MethylScope Array contains over 1 million spots each containing a unique 50 bp DNA sequence, together representing the human genome. Fluorescently labeled unmethylated and total genomic DNA differentially hybridizes to the array, resulting in a high-resolution quantitative methylation profile.
“Finding causative methylation marks is only the first step in the process,” says Jared Ordway, Ph.D., vp of research and development. “We can continue their translation into clinical diagnostics by developing a specific assessment of the methylation density of that marker. But sometimes it makes more sense to measure the downstream result.”
With a collaborator, the company is conducting clinical validation of a predictive biomarker for increased risk of colorectal cancer. Forty percent of colon tumors have loss of imprinting (silencing) of the maternal copy of IGF2, insulin-like growth factor II gene. Expression of both copies of this gene is strongly linked with colon and 20 other cancer types. And while the appearance of this biomarker is caused by epigenetic changes, the clinical assay measures the allele-specific expression of IGF2.
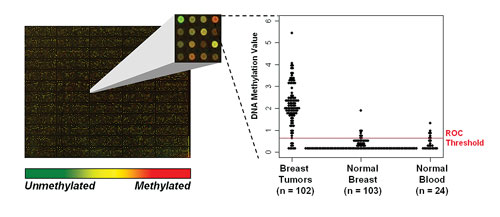
Orion Genomics’ MethylScope technology simultaneously measures the DNA methylation state of over 1 million loci across the human genome (left panel). MethylScreen, a quantitative PCR-based assay, provides precise measurements of DNA methylation content at specific sites in the genome. For example, hypermethylation of the GSHR gene, identified by MethylScope profiling and monitored in hundreds of samples by a MethylScreen assay, occurs in approximately 90% of breast tumors (right panel).
Monoclonal Antibodies
Detection of circulating blood biomarkers is commonly accomplished by sandwich ELISA using one antibody for capture and another antibody for detection. But what if that biomarker is also a target for a therapeutic monoclonal antibody (mAb)?
In a clinical sample, such a biomarker would exist in both bound and unbound forms in the presence of potentially higher amounts of the therapeutic mAb. Moreover, the binding of the therapeutic mAb to the target may change the body’s natural clearance, thereby altering the biomarker concentration in response to treatment.
Quantification of both bound and unbound biomarkers would produce important data to not only show efficacy of the mAb, but to potentially facilitate establishment of dosing levels when used in preclinical or clinical settings.
“Development of a high-throughput assay detecting a biomarker independent of the presence of a therapeutic mAb is challenging,” says Jana Chain, staff scientist at the laboratory for experimental medicine at Eli Lilly and Company.
In response to this challenge, Lilly has developed an immunoassay for quantifying total biomarker in the presence of high levels of mAb targeting the biomarker of interest. The assay utilizes a capture antibody to a noncompeting, mid-region epitope. First, the bound and unbound biomarkers are captured on a plate coated with the noncompeting antibody. Next, the biomarker is eluted with acid, then transferred and bound to a second plate. A biotinylated version of the same noncompeting antibody is then utilized for detection.
This novel format allows for quantitation of ng/mL levels of biomarkers in the presence of ug/mL levels of therapeutic mAbs. To date, Lilly has validated this assay format for three therapeutic monoclonal antibodies currently in clinical trials. “Developing an assay that enables us to quickly and efficiently screen clinical trial samples for efficacy of our therapeutic proteins is in line with our company’s strategy to improve outcomes for individual patients,” says Chain.