May 15, 2010 (Vol. 30, No. 10)
Technique’s Considerable Power Has Led to Extensive Use in Research Laboratories and Clinical Settings
Early efforts to examine genomic changes in the clinical setting relied on cytogenetic techniques such as chromosome karyotyping, a widely used approach to examine chromosomes and identify changes that could be responsible for a specific phenotype. This technique is sometimes inadequate, however, due to its crude limit of resolution, five to ten million bases, after which chromosome changes can be difficult to detect under a microscope.
An approach that has emerged over recent years, array-based comparative genomic hybridization, or array CGH, allows genome-wide scans to detect chromosomal rearrangements at a greatly improved resolution, down to hundreds of bases, or more than one thousand-fold better than karyotyping.
In array CGH, the control or reference genome and the patient’s genome are differentially fluorescently labeled and competitively hybridized to several hundreds or thousands of DNA targets. “The high resolution has become a very powerful aspect of this technology, and many times when we cannot detect any abnormalities at the chromosome level, we discover a lot of changes at the genome level by array CGH,” explains Jun Gu, M.D., Ph.D., instructor/education coordinator in the cytogenetic technology program at University of Texas and M.D. Anderson Cancer Center.
At the Association of Genetic Technologists’ annual meeting to be held next month in Phoenix, Dr. Gu will provide an update on advances at the frontline of array CGH. He will also offer a case study describing applications that are suitable for this type of analysis.
“Array CGH has emerged as a high-resolution whole-genome approach that could potentially help us classify genetic diseases with greater precision. This is the beginning of a new era, and many exciting discoveries are in front of us.”
Autism
As part of their ongoing efforts to find de novo genomic changes linked to autism, Simon G. Gregory, Ph.D., associate professor of medical genetics at Duke University, together with collaborators, used array CGH to examine the genomic profiles of 119 children with idiopathic autism.
This approach unveiled over 100 copy number variants, from which the investigators focused on a 0.7-Mb deletion on the short arm of chromosome 3, a region containing five genes. One of these genes, OXTR, encodes the oxytocin receptor that, having previously been implicated in autism, emerged as an interesting candidate to pursue.
The authors found that one child’s mother also carried the deletion, and she had self-reported obsessive-compulsive disorder symptoms, a condition previously linked to the same gene. However, to the investigators’ initial surprise, the child’s brother, despite being affected with autism, did not harbor this deletion. “We were briefly disappointed, until we decided to look at the methylation profile of this receptor,” recalls Dr. Gregory.
Previous reports documented that the OXTR gene is regulated by its DNA methylation status. By performing disulphide sequencing, Dr. Gregory and colleagues identified five CpG dinucleotides that exhibited differential methylation patterns among family members.
At three of these sites, the affected brother showed extensive methylation, an epigenetic modification correlated with decreased gene expression. Furthermore, on small groups of patients with autism, the authors showed that several of these dinucleotides were hypermethylated in peripheral blood mononuclear cells and in the temporal lobe cortex, as compared to unaffected individuals, a finding that they are proposing to further explore on larger patient groups.
Another research effort that Dr. Gregory is conducting in collaboration with medical oncologist Andrew J. Armstrong, M.D., ScM, proposes to profile copy number changes in primary and secondary malignant prostate tumor cells.
Cells that originate from primary tumors often metastasize to distant sites in various organs and establish secondary tumors that contribute to the lethal behavior of cancer; however, the factors that shape this process, and the specific affinity that primary prostate cancer cells have for certain secondary dissemination sites, are insufficiently understood.
“We are trying to identify the circulating tumor cells, pool them, and use unbiased genomic amplification to conduct copy number profiling and unveil potential copy number variations that would make primary tumor cells more likely to metastasize or increase their affinity for specific secondary tissues,” explains Dr. Gregory.
In addition, as part of a project seeking to identify factors associated with tumor behavior, Dr. Gregory and collaborators are using array CGH to explore correlations between copy number variation and the progression of oligodendroglyomas.
In addition to the research world, array CGH is also becoming commonplace in the clinical community and is increasingly replacing G-banded karyotyping as the gold standard for cytogenetic analysis.

Researchers from Duke University are using array CGH in their efforts to find de novo genomic changes linked to autism. This schematic represents the characterization of genomic rearrangements using a genomic microarray.
Task Force
A large project is under way to optimize and standardize array design and interpretation guidelines for clinical labs worldwide. This project was initiated in 2008 primarily by Emory University but also involved GeneDx, ARUP Laboratories, and Mayo Clinic. Known as the International Standard Cytogenomic Array Consortium (ISCA), this collaborative undertaking has now expanded and includes over 100 labs worldwide.
“I think this collaborative effort is necessary for the future of array CGH testing in clinical laboratories to ensure its proper use and the necessary education that goes with it,” explains Swaroop Aradhya, Ph.D., director of clinical cytogenetics and vp at GeneDx. In fact, in what amounts to a paradigm shift, ISCA is releasing a consensus statement promoting the use of array CGH as a first-tier test in children with developmental delay, intellectual disability, and/or congenital anomalies.
Array CGH has also been used outside of the cytogenetics context and in the realm of single-gene Mendelian disorder testing. At the American College of Medical Genetics’ recent “Clinical Genetics” meeting, Dr. Aradhya and his colleagues described an adaptation of the array CGH approach to evaluate individual exons of single genes each associated with a specific genetic disorder.
The GeneDx exon array can examine more than 560 genes and has now been used to test more than 2,500 patients, Dr. Aradhya says. In approximately 3.5% of these patients, this approach revealed mutations that had either eluded detection or were not identifiable with other methods.
GeneDx, which started offering cytogenetic postnatal array CGH three years ago, has expanded its array CGH service to offer prenatal cytogenetic testing for fetuses with ultrasound abnormalities, exon-level copy number analysis for single-gene disorders, and mitochondrial genome testing. Its parent company, BioReference Laboratories, also offers array CGH testing for leukemias. A few other clinical laboratories in the U.S., including Emory University and Baylor College of Medicine, now offer a similar breadth of array CGH testing services.
“Our main effort is to put together a large consortium of clinical diagnostic labs using whole-genome array technology and pool the data into a central public database that will provide a valuable resource for the research and clinical communities,” says David H. Ledbetter, Ph.D., the Robert W. Woodruff professor of human genetics and director of the division of medical genetics at Emory University.
ISCA makes the information deposited in this massive database available for vendors, registered investigators conducting research on copy number variants, and clinical laboratories. “We expect to have data from 200,000 array CGH patient samples in the next two years,” predicts Dr. Ledbetter.
The data will help compare findings from individual patients with results already deposited in the database and will help investigators during efforts to distinguish copy number variants that are benign from the ones that have pathogenic effects. “The major problem right now is that we are not sure about the functional consequence for most copy number variants, but large datasets from control and patient populations should enable us to quickly determine which ones have clinical significance.”
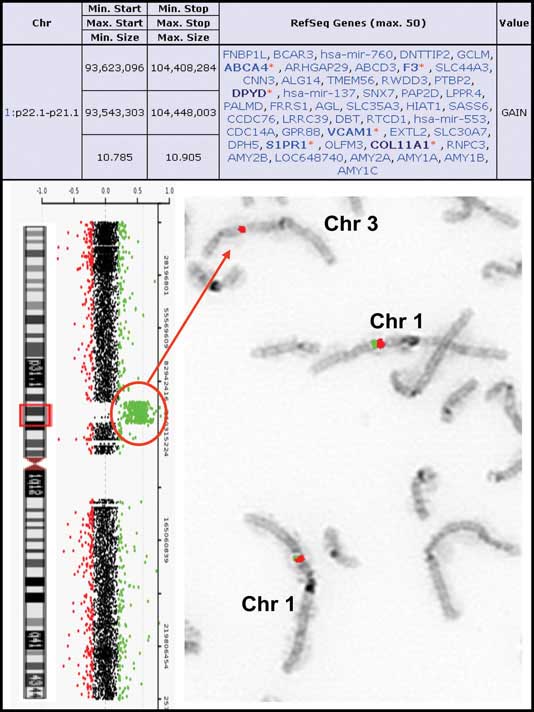
A simple insertional translocation: An aCGH summary table indicating the size of the duplication segment is on the top. To the left are the aCGH results showing the oligonucleotide probes specific for chromosome 1 next to the chromosome 1 ideogram. The probes detecting a gain in copy number are shown in the red circle. To the right is the reversed FISH image showing insertion of chromosome 1p21.3 material (probe RP11-465K1 in red) into chromosome 3q. The control probe is RP11-488L9, labeled in green. [Baylor College of Medicine]
Genetic Conditions
Dr. Ledbetter’s group is examining the contribution of rare de novo genetic changes to autism and other neurodevelopmental and psychiatric conditions. They have found clinically relevant copy number variations, most often de novo events, in about 10% of patients with autism, a result that confirms earlier findings reported by other investigators.
Routine chromosome analysis is particularly limited in its ability to detect submicroscopic microduplications and microdeletions, chromosomal rearrangements that represent the genetic basis of many medical conditions, and these chromosomal changes were shown, by several authors, to be underdetected and under-diagnosed.
“For clinicians and families, the development of CGH arrays has revolutionized the recognition of all these variants,” says Sau Wai Cheung, Ph.D., professor and director of the cytogenetics laboratory at Baylor College of Medicine.
Dr. Cheung and collaborators used high-resolution array CGH to explore the genomic basis of Phelan-McDermid or 22q13.3 deletion syndrome, a rare genetic condition characterized by neurodevelopmental delay, delayed speech, hypotonia, and minor dysmorphic craniofacial features.
On a group of 10 patients, the authors characterized the boundaries of the deletions associated with this condition and reported heterogeneous pathological rearrangements that deleted between 3 and approximately 90 genes. This result illustrated the utility of array CGH in defining the extent of deletions and characterizing their breakpoints and highlighted their important clinical applications in unveiling genotype-phenotype correlations.
In another recent study, Dr. Cheung and colleagues demonstrated the strength conferred by the combined use of array CGH and fluorescence in situ hybridization when exploring insertional translocations, a type of complex genomic rearrangement requiring at least three chromosomal breaks.
By using this combined approach to analyze samples from 18,000 patients, the authors unveiled 40 insertional translocations and reported that four of them, which resulted in deletions and most likely had pathogenic significance, were inherited from a parental balanced insertion. This finding drastically changed the implications for estimating the recurrence risk and for conducting genetic counseling in the respective families.
While new genome-wide association studies are reported on a monthly basis, understanding their significance could open major difficulties for investigators. “We are at exciting times, but there are a lot of things we don’t know. While array CGH will be here for a while, the real challenge is how to interpret the information that is generated, and this will represent the bottleneck,” predicts Dr. Cheung.
From its establishment as a field in 1956, human cytogenetics has witnessed profound transformations, first through the introduction of chromosome banding, and subsequently with the development of radioactive and fluorescence in situ hybridization, followed by comparative genomic hybridization.
As one of the most recent and sophisticated additions to this set of tools, array CGH promises to unveil a previously undetectable facet of chromosome organization. This new level of scrutiny becomes a testimony of the recondite mysteries that biological systems still have to offer, so vividly illustrated in Albert Einstein’s words: “We still do not know one thousandth of one percent of what Nature has revealed to us.”
A simple insertional translocation: An aCGH summary table indicating the size of the duplication segment is on the top. To the left are the aCGH results showing the oligonucleotide probes specific for chromosome 1 next to the chromosome 1 ideogram. The probes detecting a gain in copy number are shown in the red circle. To the right is the reversed FISH image showing insertion of chromosome 1p21.3 material (probe RP11-465K1 in red) into chromosome 3q. The control probe is RP11-488L9, labeled in green.[Baylor College of Medicine]



