Beste Calimlioglu Department of Bioengineering, Marmara University and Istanbul Medeniyet University
Kubra Karagoz Department of Bioengineering Marmara University
Tuba Sevimoglu Department of Bioengineering Marmara University
Elif Kilic Department of Bioengineering Marmara University
Esra Gov Department of Bioengineering Marmara University
Kazim Yalcin Arga Department of Bioengineering Marmara University
An Integrative Analysis of Transcriptomics and Protein–Protein Interaction Data
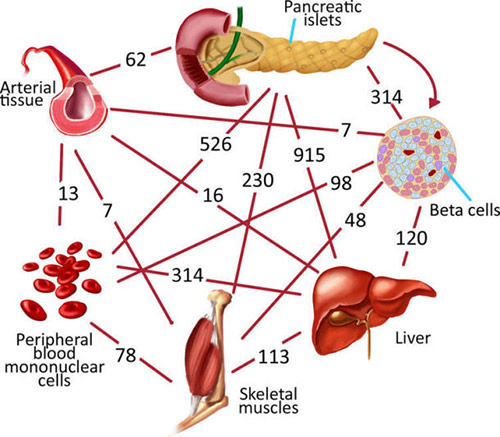
Type 2 diabetes mellitus (T2D) is a major global health burden. A complex metabolic disease, type 2 diabetes affects multiple different tissues, demanding a ‘‘systems medicine’’ approach to biomarker and novel diagnostic discovery, not to mention data integration across omics-es (Günther et al. 2014; Montague et al. 2014; Sahu et al. 2014). The two important key determinants of T2D are the failure of peripheral tissues (such as liver, muscle, and adipose tissue) to respond to insulin doses (so-called insulin resistance), and the failure of suitable insulin secretion by pancreatic betacells in response to increased blood glucose levels (Kaiser and Oetjen, 2014).
The duration of hyperglycemia caused by failure of betacells also affects insulin secretory capacity, mass, and apoptosis rate of beta-cells, resulting in additional alterations in several processes such as islet inflammation, amyloid deposition, critical B-cell phenotypic alterations (Prentki and Nolan, 2006). On the other hand, the state of hyperglycemia damages nerves and blood vessels, leading to major healthrelated issues such as cardiovascular diseases, stroke, blindness, dental problems, and diabetes-related amputations. Other complications of T2D include enhanced vulnerability to neurodegenerative diseases, presence of various cancer types, pregnancy problems, loss of mobility with aging, and depression (Musselman et al., 2003; Retnakaran et al., 2006).
Due to the high prevalence of T2D and its fateful complications, identifying the genes or genetic factors associated with the development of T2D and elucidating the mechanisms underlying the disease are crucial in prognosis, and development of personalized medicine and therapeutic strategies.
Since it is a polygenic disorder (i.e., multiple genes located on different chromosomes take active roles in the development of the disease), it is better to reveal that gene expression varies more across tissues than across individuals. Several studies reported findings on T2D gene expression profiles of
different tissues individually (Kazier et al., 2007; Cangemi et al., 2011; Misu et al., 2010; van Tienen et al., 2012; Dominguez et al., 2011). Despite individual studies exploring T2D specific genes in various tissues, studies considering the meta-analysis of diverse transcriptomics datasets and integrating gene expression profiles with biological networks are very limited.
Keller and co-workers (2008) studied gene expression profiles in eight experimental groups of lean and obese mice.
To read the rest of this article click here.
OMICS: A Journal of Integrative Biology integrates global high-throughput and systems approaches to 21st century science from “cell to society” – seen from a post-genomics perspective. The above article was first published in the September 2015 issue of OMICS: A Journal of Integrative Biology with the title “Tissue-Specific Molecular Biomarker Signatures of Type 2 Diabetes: An Integrative Analysis of Transcriptomics and Protein–Protein Interaction Data”. The views expressed here are those of the authors and are not necessarily those of OMICS: A Journal of Integrative Biology, Mary Ann Liebert, Inc., publishers, or their affiliates. No endorsement of any entity or technology is implied.