This movie shows the helicase protein complex from all angles, and reveals how its shape changes back and forth between two forms. The research team hypothesizes that the rocking action of this conformational change could help split the DNA double helix and move the helicase along one strand so it can be copied by DNA polymerase.
![These are two images showing the structure of the helicase protein complex from above. (a) A surface-rendered three-dimensional electron density map as obtained by cryo-EM. (b) A computer-generated 'ribbon diagram' of the atomic model built based on the density map. The helicase has three major components: the Mcm2-7 hexamer ring in green, which encircles the DNA strand; the Cdc45 protein in magenta; and the GINS 4-protein complex in marine blue. Cdc45 and GINS recruit and tether other replisome components to the helicase, including the DNA polymerases that copy each strand of the DNA. [Brookhaven National Laboratory]](https://genengnews.com/wp-content/uploads/2018/08/108310_web1851822472-1.jpg)
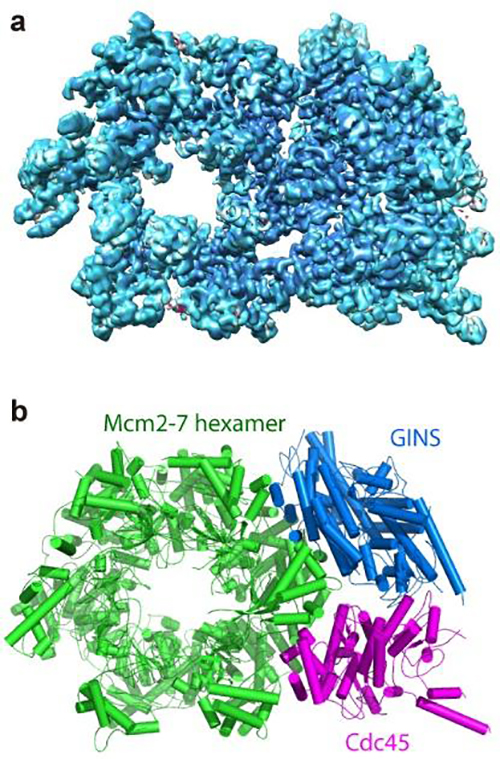
These are two images showing the structure of the helicase protein complex from above. (a) A surface-rendered three-dimensional electron density map as obtained by cryo-EM. (b) A computer-generated ‘ribbon diagram’ of the atomic model built based on the density map. The helicase has three major components: the Mcm2-7 hexamer ring in green, which encircles the DNA strand; the Cdc45 protein in magenta; and the GINS 4-protein complex in marine blue. Cdc45 and GINS recruit and tether other replisome components to the helicase, including the DNA polymerases that copy each strand of the DNA. [Brookhaven National Laboratory]
A collaborative team of researchers from the U.S. Department of Energy's Brookhaven National Laboratory, Stony Brook University, Rockefeller University, and the University of Texas have just released detailed structural images of DNA helicase from yeast and are proposing a novel mechanism for how the molecular machinery functions. The scientists believe their new data could provide valuable insight into ways that DNA replication can go askew.
“DNA replication is a major source of errors that can lead to cancer,” explained senior study author Huilin Li, Ph.D., a professor with a joint appointment at Brookhaven Lab and Stony Brook University. “The entire genome, all 46 chromosomes, gets replicated every few hours in dividing human cells, so studying the details of how this process works may help us understand how errors occur.”
The findings from this study were published recently in Nature Structural & Molecular Biology through an article entitled “Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation.”
This current study builds upon previous work that produced the first-ever images of the complete DNA-copying protein complex, called the replisome. That study provided a surprising revelation about the location of the DNA-copying enzymes, DNA polymerases. This new study focuses on the atomic-level details for the helicase portion of the protein complex—the part that encircles and splits the DNA double helix so the polymerases can synthesize two new daughter strands.
As they had done in their previous work, Dr. Li and his colleagues produced high-resolution images of the helicase using cryo-electron microscopy (cryo-EM). This technique holds an advantage over other EM methods in that proteins can be studied in solution, closely replicating intracellular conditions.
“You don't have to produce crystals that would lock the proteins in one position,” Dr. Li remarked. It's an important point because helicase is a molecular machine made of 11 associated proteins that must be flexible to work. “You have to be able to see how the molecule moves to understand its function,” Dr. Li said.
Once the images of the replication machinery were assembled, the investigators were able to map out the locations of the individual amino acids that make up the helicase complex in each conformation. Then, combining those maps with existing biochemical knowledge, they came up with a mechanism for how the helicase works.
“One part binds and releases energy from a molecule called ATP. It converts the chemical energy into a mechanical force that changes the shape of the helicase,” Dr. Li stated. The molecule subsequently ejects the drained ATP and the helicase complex reverts to its original shape so a new ATP molecule can come in and begin the process again.
“It looks and operates similar to an old-style pumpjack oil rig, with one part of the protein complex forming a stable platform, and another part rocking back and forth,” Dr. Li noted. The researchers postulate that the rocking motion would nudge the DNA strands apart and move the helicase along the double helix in a linear fashion.
This direct translocation mechanism appears to be quite different from the way helicases are thought to operate in more primitive organisms such as bacteria, where the entire complex is believed to rotate around the DNA. However, there is biochemical evidence to support the idea of linear motion, including the fact that the helicase can still function even when the ATP hydrolysis activity of some, but not all, of the components is knocked out by mutation.
“We acknowledge that this proposal may be controversial, and it is not really proven at this point, but the structure gives an indication of how this protein complex works, and we are trying to make sense of it,” Dr. Li stated.