September 1, 2015 (Vol. 35, No. 15)
Chromatin Factors Pretty Much Allow Us to Live Out Of the Nuclear Suitcase
Chromatin-associated factors play an important role in the epigenetic regulation of gene expression. These factors include various proteins that contribute to chemical alteration of DNA or histones, as well as those that control chromatin structure at the nucleosome level.
Extensive studies have documented chromosomal aberrations and dysregulation of chromatin factor gene expression in many types of cancer. A single defect in a chromatin factor can potentially affect the expression patterns of several hundred target genes, resulting in the disruption of multiple cellular pathways.
As further research into chromatin factors provides an increasing level of detail regarding their interactions with multiple targets, cancer epigenetics is emerging as a rapidly developing area of research. As a result, the dysregulation of chromatin factor gene expression offers attractive opportunities for the development of therapeutic agents.
The topic will be highlighted at Chromatin and Epigenetics in Cancer, an American Association for Cancer Research (AACR) conference that will be held September 24–27 in Atlanta. Cancer epigenetics has also been discussed at meetings that were more application oriented, such as CHI’s Epigenetic Inhibitor Discovery conference held earlier this year in San Diego.
“Histone modifications can alter chromatin in two ways,” says Catherine A. Musselman, Ph.D., assistant professor of biochemistry at the University of Iowa, Carver College of Medicine. “They may directly alter chromatin contacts, or they may indirectly remodel the structure.”
Direct mechanisms involve changes at histone-histone or histone-DNA contacts that affect chromatin conformation. One common modification is acetylation of lysine residues in histones, which neutralizes their positive charge and disrupts their electrostatic interactions with other proteins. Indirect effects involve chromatin remodeling cofactors, whose interaction with specifically modified histones can recruit them to certain regions of chromatin, for example, during the transcription process.
Dr. Musselman’s group studies histone lysine methylation, one of the most diverse histone modifications. It can have a variety of functional effects. “For instance, trimethylation of histone H3 at Lys4 (H3K4me3) is a marker of promoters of transcriptionally active genes,” notes Dr. Musselman. In contrast, trimethylation of lysine 27 (H3K27me3) is typically associated with gene repression.
In particular, Dr. Musselman’s research focuses on a class of proteins known as plant homeodomain (PHD) finger proteins. PHD finger protein 1 (PHF1) is an essential component of nuclear protein complexes like the transcriptional repressor polycomb repressive complex 2 (PRC2), as well as the DNA damage repair enzymes Ku70/Ku80.
“Dysregulation of PHF1 alters global H3K27me3 levels, which leads to altered gene expression levels,” explains Dr. Musselman. The exact effects are gene-dependent; however, the importance of PHF1 to the DNA damage response pathway, as well as its reported involvement in p53-mediated pathways, suggest that it has multiple roles in tumorigenesis.

Researchers at the University of Iowa studied how PHD finger protein 1 (PHF1) binds to histone H3 trime- thylated at Lys36 (H3K36me3). They found that PHF1’s Tudor domain (light purple) binds to the histone’s methylated tail (pink). Note that the aromatic residues of the Tudor domain that constitute the methyl-lysine binding pocket are highlighted in blue. [Image courtesy of Catherine A. Musselman, Ph.D.]
Interactions with RNA Polymerase II
Brian Strahl, Ph.D., a professor of biochemistry and biophysics at the University of North Carolina School of Medicine, is addressing several intriguing issues that surround the mechanisms by which distinct chromatin structures are established and maintained, as well as how the underlying DNA is made accessible to the transcriptional machinery. Much of his research centers on the properties of the yeast transcription factor Set2 and the human equivalent, SETD2.
“Set2/SETD2 is a histone methyltransferase I first identified as a postdoctoral fellow,” says Dr. Strahl. At the time, little was known about the role of histone methylases in chromatin function. “By tethering Set2 to gene promoters, I was able to initially show that Set2 and its methylation in yeast cells had the ability to turn off transcription,” he adds.
Although Set2 has been extensively studied in yeast, the role of SETD2 in humans is less clear. Like Set2, SETD2 appears to associate with RNA polymerase II during transcription to trimethylate the lysine-36 residue in histone H3 (H3K36me3). This process is a key factor in the response to double-stranded breaks (DSBs) in DNA, a phenomenon observed in multiple types of cancer. SETD2 is one of the most frequently mutated proteins in cancer. “The frequency of mutation is especially high in bladder, breast, gliomas and most significantly, kidney cells,” explains Dr. Strahl.
The question of how RNA polymerase II senses and responds to DNA damage in cancer remains unanswered. One possibility is that it pauses at the site of DSBs. “At that point,” notes Dr. Strahl, “repair machinery coupled with the polymerase, along with DNA damage response machinery, would then take action.” Future research in Dr. Strahl’s laboratory will explore the question of how SETD2 loss of function may result in inappropriately repaired DNA and genomic instability—a characteristic of many cancer types.
Jumonji Family of Demethylases
Histone methylation of lysine and arginine residues, especially trimethylation, was long considered to be irreversible. However, recent research has shown that the “erasing” of histone methyl marks by a group of demethylase enzymes also plays a significant role in epigenetic regulation of gene transcription.
Kristian Helin, Ph.D., director and professor at the Biotech Research & Innovation Centre (BRIC), University of Copenhagen, studies a class of demethylases known as Jumonji (JmjC) proteins. “JmjC-containing histone demethylases can reverse the effect of histone methyltransferases, but in general they do not appear to be regulators of transcription,” says Dr. Helin. Instead, these proteins stabilize the transcriptional complexes formed by transcription factors and other proteins involved in cell signaling pathways.
One member of the JmjC family, UTX, is a demethylase specific to H3K27me3/me2 modifications. In vivo, UTX cooperates with the histone methyltransferase MLL4; these proteins are part of a complex that act as a transcriptional activator. Mutations in the gene encoding UTX are associated with “many different types of cancer including renal cell carcinomas, leukemia, and other hematopoietic cancers,” according to Dr. Helin.
Future research in Dr. Helin’s group will use genetically engineered mouse models to assess the roles of specific histone-modifying enzymes in cancer. The goal, declares Dr. Helin, is “elucidation of how these enzymes contribute to normal proliferation and development, and how their deregulation leads to cancer.”
Inhibitors of Jumonji Enzyme Activity
JmjC demethylases also constitute the primary area of research for Elisabeth D. Martinez, Ph.D., an assistant professor of pharmacology at the University of Texas Southwestern Medical Center. “In cancer,” says Dr. Martinez, “the expression of some histone demethylases is highly increased, and this hyperactivity leads to aberrant gene expression and defects in the normal regulation of growth pathways.”
Dr. Martinez is investigating the therapeutic potential of a small molecule drug candidate known as JIB-04. “In biochemical assays using recombinant Jumonji enzymes, JIB-04 inhibits their histone demethylase activity,” explains Dr. Martinez. Her studies also showed that JIB-04 inhibited histone demethylase activity on H3K9me3 in lysates of cancer cells but not in patient-matched normal cells.
While further studies will provide mechanistic details, Dr. Martinez suggests that “JIB-04 blocks tumor growth and prolongs survival by reprogramming the tumor’s transcriptional profile,” and that the process is mediated, in part, by the drug’s effects on Jumonji demethylase activity.
Other researchers have studied different classes of histone demethylases inhibitors, including alpha-ketoglutarate analogs. Compared to these compounds, however, JIB-04 is cell-permeable and has better efficacy in the mouse models tested. “In addition, data to date demonstrate that it is markedly more specific than alpha-ketoglutarate analogs and inhibits Jumonji demethylases but not other cellular alpha-ketoglutarate–dependent enzymes,” observes Dr. Martinez.
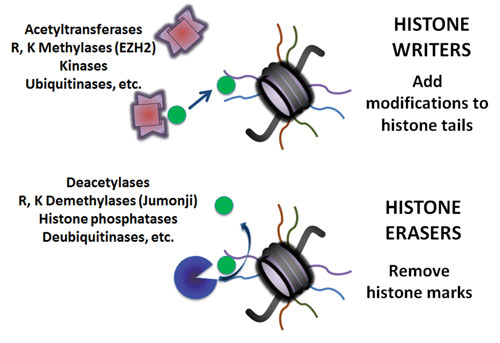
At the University of Texas Southwestern Medical Center, researchers focus on the role of Jumonji histone demethylases in cancer. This image, prepared by Juan Bayo-Fina, Ph.D., depicts various means by which histone epigenetic marks may be added or removed. [Image courtesy of the Martinez Lab]
Clinical Trials
Several companies are targeting histone modification pathways in efforts to bring new drugs to the market for cancer therapy. One such company is Cambridge-based Constellation Pharmaceuticals. The company recently announced the initiation of a Phase I trial of a compound known as CPI-1205, which targets pathways mediated by the protein Enhancer of Zeste Homolog 2 (EZH2).
“EZH2 functions as the catalytic component of the multisubunit complex PRC2,” says Robert Sims, executive director of biology at Constellation. “This complex functions in the maintenance of gene silencing, in part due to the ability of EZH2 to methylate H3K27.”
Sims explains that mutations in the EZH2 gene that alter its substrate specificity have been associated with several non-Hodgkin lymphoma subtypes (including diffuse large B-cell lymphoma and follicular lymphoma) and melanoma. He adds that “CPI-1205 inhibits the EZH2 methyltransferase activity, and thus impedes EZH2’s potential to repress gene expression in lymphoma.”
Constellation is also evaluating other inhibitors, including one that targets the bromodomain and extra-C terminal domain (BET) family of chromatin adaptors. These proteins “bridge” transcriptional coactivators to acetylated histones during a late stage of the transcription cycle, typically at transcriptional pausing of RNA polymerase II. Dysregulation of BET gene expression is linked to NUT midline carcinoma, a rare, aggressive epithelial cancer characterized by a chromosomal translocation in the nuclear protein in testis (NUT) gene.
“Many additional cancers utilize BET proteins to drive oncogenic gene expression, such as MYC, MYB, and BCL-2,” notes Sims. These cancers include leukemia, lymphoma, multiple myeloma, and neuroblastoma.
Constellation is actively pursuing research in other chromatin-modifying pathways, including reversible lysine methylation and acetylation. “This includes all aspects of these signaling pathways—writing, erasing, and reading,” asserts Sims. He also indicates that the company is exploring other aspects of chromatin structure and function, with an emphasis on modulating gene regulatory circuits.



