December 1, 2017 (Vol. 37, No. 21)
Synthetic Biology Is Enabling Optimization that Is Iterative—and Remunerative
At the dawn of the scientific revolution, the idea that humans could improve on nature was deemed foolish, if not blasphemous. It was even said that to paint the lily, or to throw perfume on the violet, or to add another color to the rainbow, would be “wasteful and ridiculous excess.” Now we know better.
Nature, as represented by microbial organisms, is up for human-engineered improvement—and not just the sort of improvement that can be achieved by mere tinkering. Craftwork in the modification of microbial organisms is fast giving way to industrial-design approaches. Microbial organisms are being stripped down and streamlined so they can be equipped with new modules and “tricked out.”
These approaches are being implemented systematically by synthetic biology startups. A few representative startups—Synthetic Genomics, Synlogic, and Zymergen—are highlighted in this article. All of them are converting microbial organisms into bioengineering platforms that can support a design-build-test-repeat approach to product development. Each startup, however, is taking this iterative approach in a different direction.
Synthetic Genomics is developing protein-expression platforms for the biotech industry. Synlogic is producing “living medicines,” probiotics for patients with metabolic disorders or autoimmune disease. Zymergen is developing a platform that can generate specialty materials of interest to diverse industries.
These startups often task themselves with squeezing more performance out of microbial organisms that have already been optimized. In a sense, the startups are trying to “gild refined gold.” The pursuit is by no means futile. By combining biology with automation and data-science strategies, the startups are increasing product output and reducing production costs while scaling processes from the bench to the bioreactor.
Cell Lines on the Assembly Line
“When we consider what we are doing now to enhance living systems, we have to look back at history,” states Matthew Weinstock, Ph.D., a scientist at Synthetic Genomics. “In the early 1940s, producing penicillin meant taking a living system off the rack and coaxing it to produce at a larger scale. Then in the 1970s, scientists developed protocols for recombinant DNA that allowed one, two, or maybe a handful of modifications. Now we consider a bacterium as a chassis and perform large-scale engineering on it to make sweeping changes.”
Synthetic Genomics, through its SGI-DNA subsidiary, genetically engineered VmaxTM, a bacterial host organism designed for improved recombinant protein expression. The Vmax chassis is Vibrio natriegens, a bacterium initially isolated from salt marsh mud in Sapelo Island, GA. It was originally classified as Pseudomonas natriegens, but after it was characterized further, it was recognized as a facultative anaerobe and reassigned to the genus Vibrio.
V. natriegens fit the bill for Synthetic Genomics because it is nonpathogenic and the fastest growing bacterium known. Some strains have a doubling time of about 10 minutes, a figure indicating that this bacterium can reproduce two to three times faster than Escherichia coli, the traditional go-to bioproduction workhorse (Figure 1). V. natriegens is superior to E. coli in other respects: It grows to a higher density in culture, it produces more biomass, and it uses cheaper carbon sources to fuel its growth.
All of these features, notes Dr. Weinstock, can “help drive down production costs.” Vmax is compatible with most common expression plasmids and promoters used in E. coli for protein expression. Under normal lab conditions at 30 °C, Vmax can produce twice the biomass and four times the protein versus E. coli. That temperature also optimizes protein expression and solubility, while slightly slowing growth. In shake flasks, Vmax expresses higher levels of soluble recombinant proteins than does E. coli. “It’s not a picky eater,” states Dr. Weinstock, who points out that Vmax can grow on a wide variety of carbon sources, including sucrose.
The Synthetic Genomics scientists who engineered Vmax had to grapple with the usual bottlenecks in protein production: the need to wait for cultures to reach a high enough density to induce, compounded by the need to wait for enough cell growth to obtain sufficient biomass. To obtain greater yields of soluble protein, scientists optimized a strain of V. natriegens by removing selected pathways and inserting new synthetic DNA sequences to replace some endogenous ones:
- A highly regulated, IPTG (isopropyl β-D-1-thiogalactopyranoside)-inducible T7 RNA polymerase was introduced (analogous to the T7-expression system found in some E. coli strains), allowing for higher expression of proteins of interest.
- Nucleases and proteases were removed to maintain integrity of isolated plasmid DNA and to improve integrity of soluble protein, respectively. (“Since V. natriegens evolved to deal with harsh environmental insults, for our production purposes we removed pathways that aren’t necessary in a controlled laboratory environment in order to maximize the availability of cellular resources for our desired production goals,” states Dr. Weinstock.)
- Catalase, an enzyme that reduces hydrogen peroxide, was added to some strains to improve strain robustness for laboratory growth.
If the Vmax is used instead of E. coli, less time is spent streaking plates for sufficient colonies, inoculating cultures, and isolating plasmid DNA. Up to a day can be shaved off a production cycle run. Vmax is also compatible with E. coli expression vectors, antibiotics, and growth media. For future challenges in the field of biomanufacturing, Dr. Weinstock sees improving expression of human proteins in bacteria and yeast. “The field,” he admits, “isn’t good at that yet.”
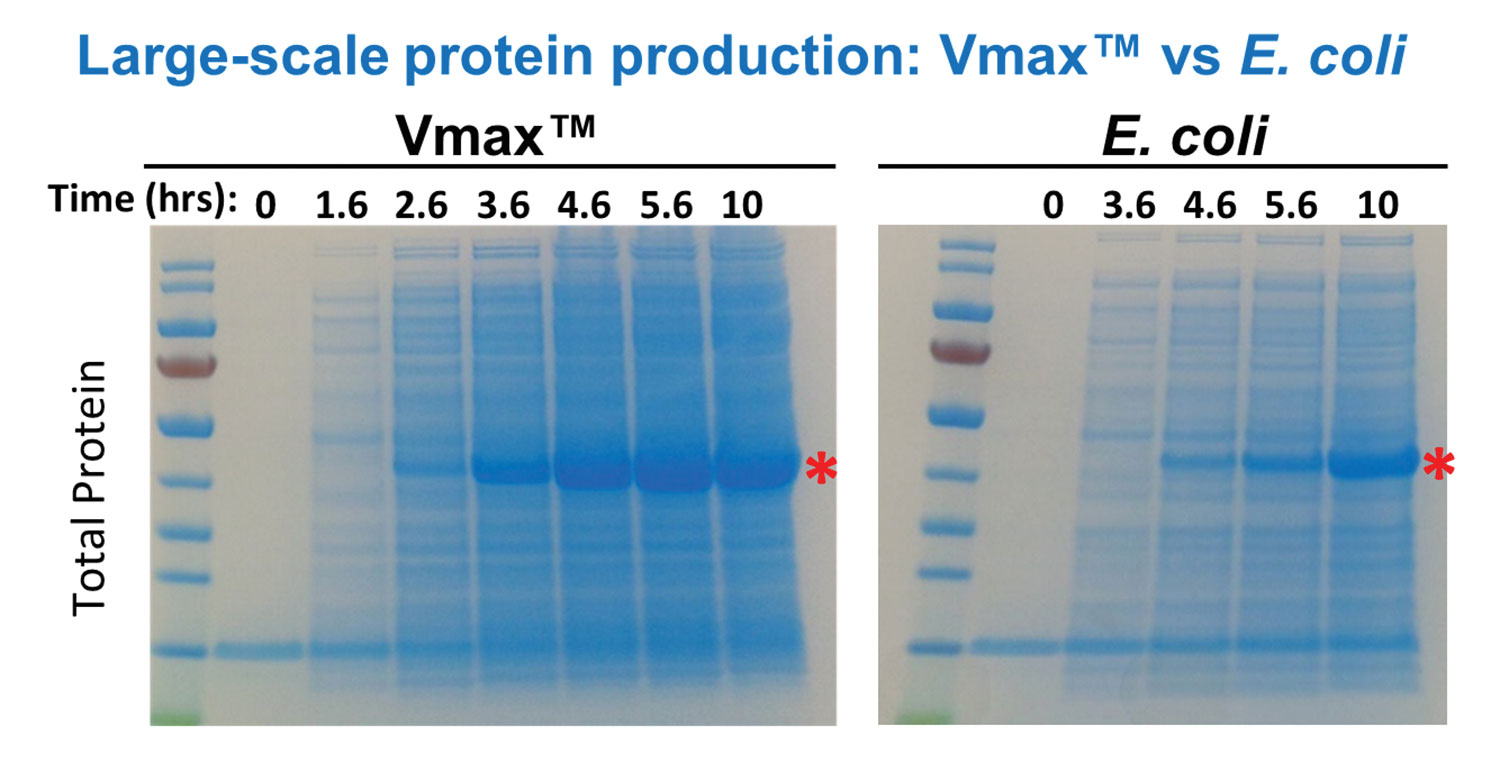
Figure 1. Vmax, a bacterial host organism designed for recombinant protein expression, outperforms E. coli-based systems, generating soluble proteins faster, more efficiently, and in larger quantities. Developed by SGI-DNA, a subsidiary of Synthetic Genomics, Vmax is derived from the marine microorganism Vibrio natriegens, a fast-growing, nonpathogenic bacterium.
Gutting It Out with Living Medicines
Synlogic scientists re-engineer genetic circuitry to enable bacteria to degrade metabolites or synthesize products more efficiently. When the re-engineered circuitry is united with E. coli Nissle, the human gut microbe that serves as Synlogic’s chassis, the result is a Synthetic Biotic™, an organism that can generate medicines that can decrease inflammation; compensate for inborn genetic errors that cause rare metabolic diseases; or treat relatively common diseases such as cancer or metabolic conditions such as diabetes and obesity.
The company’s initial clinical programs are focused on Synthetic Biotics to treat orphan metabolic diseases such as urea cycle disorders and phenylketonuria. Another Synlogic product is a synthetically engineered biotic that produces butyrate.
“High permeability in gut epithelium is correlated with a deficiency in short-chain fatty acids in the gastrointestinal tract, most notably butyrate,” says Paul Miller, Ph.D., Synlogic’s chief science officer. “Our goal is to engineer a safe, butyrate-producing probiotic to compensate for low production of natural, endogenous butyrate.”
Normally, butyrate is synthesized in the large intestine by slow-growing anaerobes such as Clostridia as a byproduct of fermentation of undigested plant fiber. Butyrate helps regulate fluid transport across gut epithelium, reduces mucosal inflammation and oxidative stress, reinforces the gut’s epithelial defense barrier, and modulates visceral sensitivity and intestinal motility. Studies have indicated that butyrate may help prevent and inhibit colorectal cancer.
Synlogic’s proprietary platform uses E. coli Nissle as its chassis for butyrate production because the organism is nonpathogenic and is easy to scale and manipulate. Discovered a century earlier by Alfred Nissle, this Gram-negative bacterium was isolated from a hospitalized German soldier who, though dispatched to a region highly contaminated with pathogenic Shigella bacteria, resisted shigellosis. He was found to possess a noticeably strong intestinal constitution.
“To re-engineer the E. coli Nissle chassis for butyrate biosynthesis, we inserted a Clostridia-derived biosynthetic operon under the control of a Tet (tetracycline)-inducible promoter into it,” states Dr. Miller. “Clostridia occur naturally in the gut and correlate with healthy gut flora.”
In butyrate’s biosynthetic pathway, this operon converts acetyl coenzyme A to butyrate as a reduced product, permitting the recycling of NADH+ to NAD. Synlogic scientists reduced the Clostridia pathway from eight genes to five by simplifying the NADH recycling process, as well as the final conversion of butyryl-coenzyme A to butyrate. The resultant pathway produced high levels of butyrate in response to activation of the Tet promoter.
E. coli uses the master transcriptional regulator FNR (fumarate and nitrate reductase) to control anaerobic gene expression. Since this regulatory mechanism could be exploited to activate gene expression in the mammalian gut’s anaerobic environment, the modified butyrate cassette was put into a low copy plasmid vector under the control of an FNR-responsive promoter.
The resulting modified strain, called SYN363, was tested for butyrate production in rich media with 0.5% glucose under both aerobic and anaerobic conditions. Under anaerobic conditions, SYN363 showed a fivefold increase in butyrate production over aerobic conditions. Further, SYN363 was tested against butyrate-producing Clostridia strains in rich media with 0.5% glucose. After a 24-hour growth period, the modified strain matched the productivity of the natural butyrate-producing strains (Figure 2). This level of biosynthetic output was demonstrated when carbon sources similar to those in the colon were available, sources such as glucoronate.
“Our driving goal was not simply to produce more butyrate per cell than we could with Clostridia, but to ensure that our product had a biological effect,” Dr. Miller states. “In preclinical models, SYN363 showed efficacy in reducing markers of inflammation in a murine model of DSS (dextran sulphate sodium)-induced colitis. Two markers of inflammation and barrier function were suppressed using SYN363.”
In anticipation of future clinical studies, the Synlogic scientists stably integrated the engineered cassette for the circuit into the E. coli Nissle genome. This cassette was shown to produce butyrate at levels comparable to those produced by the plasmid version of the circuit. To evaluate the strain further, Synlogic is working with AbbVie. The companies have been collaborating to investigate treatments for inflammatory bowel disease.
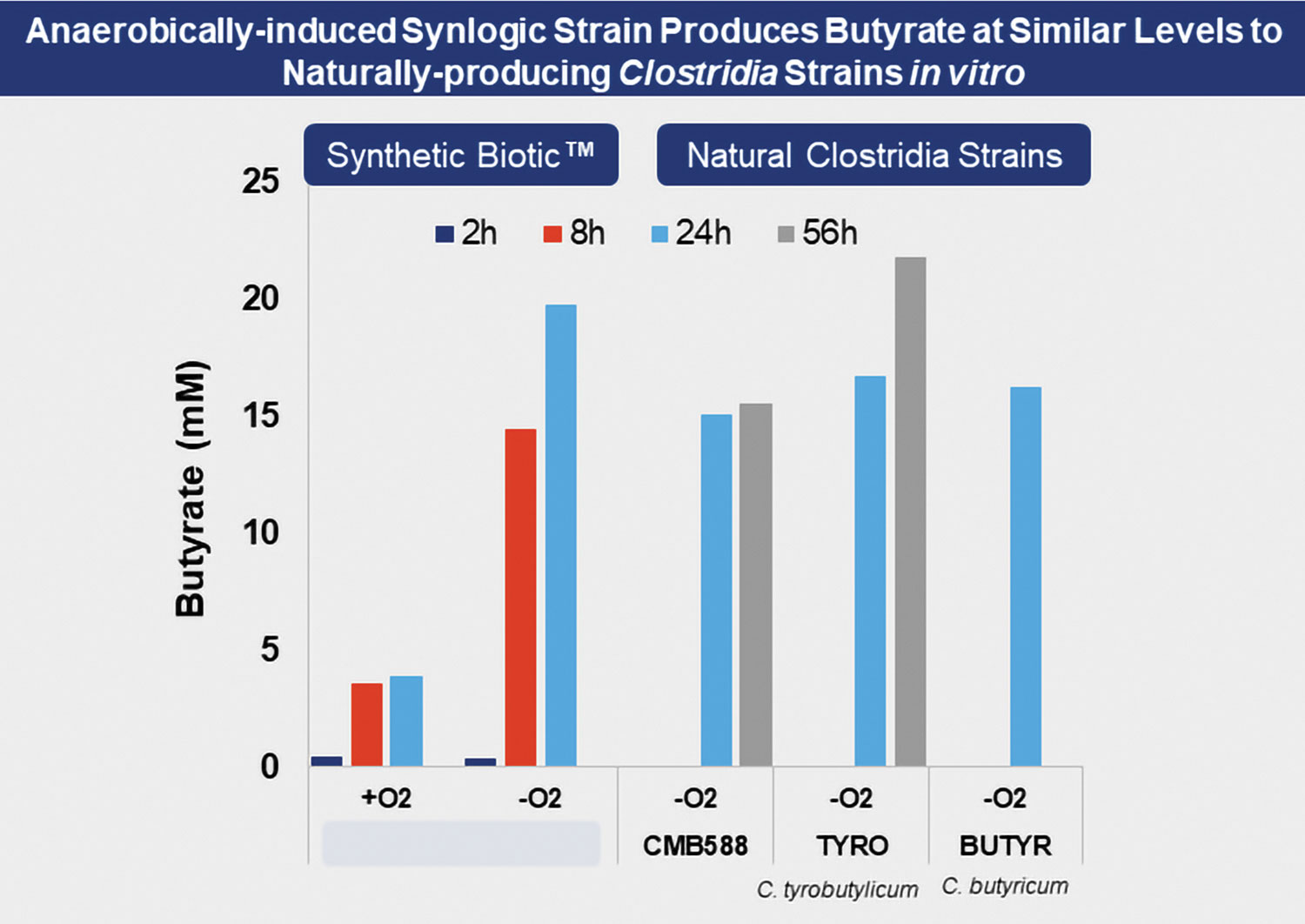
Figure 2. A probiotic bacterial strain was engineered by Synlogic to produce butyrate, a short-chain fatty acid that fuels epithelial colonocytes, drives regulatory T-cell differentiation, and helps maintain colonic mucosal health. The engineered strain, which Synlogic calls a Synthetic Biotic, rivals the butyrate production of natural Clostridia strains in vitro.
Machines of Iterative Grace
Synthetic biology may realize the utopian vision described in the Richard Brautigan poem, All Watched Over by Machines of Loving Grace, which suggested, back in 1967, that mammals and computers could live together in “mutually programming harmony.” Brautigan’s vision has always seemed far-fetched, not to mention naïve, to technology skeptics. But now that synthetic biology is on the scene, people and benevolent machines may start working together via microbial intermediaries.
Useful microbes don’t have to be designed, built, tested, analyzed, and redesigned by humans. This cycle can be managed by machines, guided by artificial intelligence or, rather, machine-learning systems. Such systems are being developed by Zymergen.
The company designs, engineers, and optimizes microbes at every point in strain development for clients’ industrial applications in agriculture, chemical and materials, and pharmaceuticals, among others. For one customer, Zymergen reduced time-to-market by more than three years; for another customer, the company more than doubled net product margin.
The company can partner with clients in three ways:
- Improve the economics of existing strains by identifying and optimizing on- and off-pathway targets.
- Help customers bring products to market faster by enabling cost-effective commercial production by optimizing known biosynthetic pathways.
- Develop novel molecules for materials not made through biology today by assembling biosynthetic pathways for new products or for products traditionally manufactured.
“We recognize pathway and genome optimization as a data and algorithms problem; automation enables us to think of it as a search function,” states Aaron Kimball, chief technology officer at Zymergen. “This search function involves two phases: exploration and exploitation.”
During exploration, it’s not obvious whether a particular genetic change will be beneficial or deleterious, so we focus on making simple changes to find areas of interest. During exploitation, subsequent iterative changes are made to optimize a particular location in the genome.
“Given finite lab capacity and client deadlines,” notes Kimball, “we’re constantly looking at data to inform how we trade-off capacity between the two [search phases].” There are also trade-offs between complexity versus number of changes.
“Microbial genomes, particularly those that are industrially relevant, are poorly understood today,” admits Kimball. “The search space is too vast to identify all the genetic changes that could elicit phenotypic improvement using traditional methods alone.
“Mechanistic understanding of cellular metabolism remains very poor, so scientists don’t know where in the genome to look for sources of improvement. And even if they did, the number of possibilities is too vast to test everything, even in an automated lab.”
Clients’ needs determine the phenotypic improvements to target and the types of assays to develop. Some clients want higher yield; others require more efficient feedstock use. “There is no one-size-fits-all approach,” insists Kimball. Different assays are developed for each cell, target product, and phenotype that provide enough predictive power to detect subtle improvements, and trade-offs are made between cost, speed, and precision.
“The ability to perform tests in high throughput lets us run more hypotheses,” Kimball continues. “High-throughput tests, however, may be less capable of picking up small signals than a slower or more expensive test.”
Factors such as epistasis are considered regardless of target phenotype. In epistasis, the (phenotypic) effect of one gene (locus) depends on that gene interacting with one or more genes at different loci.
“With epistasis, we may find multiple individual genetic sources of improvement that when combined are actually deleterious,” states Kimball. “We have developed proprietary algorithms to predict the best-performing combinations of genetic changes to avoid these negative epistatic events in product optimization. This is driven by data rather than by a mechanistic understanding of the genome.”
Optimizing the Optimized
Another important factor in product optimization is the maturity level of a cell line. “A product in the early stages of development can take advantage of certain optimization approaches that aren’t as productive in a more mature cell that is already close to its theoretical maximum target,” explains Kimball. For such a cell, the “lower-hanging fruit has been picked.”
Kimball thinks that taking a purely in silico approach is often a poor predictor of how biology behaves, and that the strategy of focusing on molecular biology on the benchtop, with software taking a backseat—while having the benefit of empiricism—lacks high throughput. “Zymergen is the ‘Goldilocks’ of these approaches—fully technology and fully biology,” asserts Kimball. “We use machine learning to predict as much as possible to inform our search function, and still test it in a wet lab.”



