October 15, 2008 (Vol. 28, No. 18)
miScript Mimics and Inhibitors Work Together to Unravel Function of miRNAs
The control of gene expression governs all life. Some of that control—maybe even a fair amount of it—depends on microRNA (miRNA). These single strands of 21–25 nucleotides come from DNA that was once called junk. Nonetheless, it is junk no more. Instead, scientists now know that miRNAs regulate the expression of genes, and appear to be involved in an ever-expanding range of biological pathways in animals, plants, and viruses.
Moreover, miRNA takes part in a wide range of today’s research. In fact, a recent search of PubMed turned up 3,266 hits for articles on these molecules. Still, research scientists know that today’s knowledge of and applications for miRNA are only the beginning of these areas of research. Consequently, getting the most from miRNA—in terms of unraveling its natural roles and finding more ways to use it in research—depends on the development of standardized miRNA-based tools.
Already, research shows that miRNAs participate in fundamental biological processes, including apoptosis, development, and differentiation. In other words, these molecules orchestrate the on-and-off patterns of genes that work from life’s beginning until its end. In addition, malfunctions in the expression of miRNA genes themselves lead to misregulation that plays some role in a range of diseases including various cancers and heart disease.
As the list of natural jobs performed by miRNA increases, the number of known miRNAs rises even faster. In mid-2007, for example, the Wellcome Trust Sanger Institute’s miRBase consisted of just over 5,000 miRNAs. By April 2008, when Sanger brought out Release 11.0, it contained nearly 6,400 miRNAs.
Given the emergent nature of miRNA-based studies, the research often runs ahead of the technology that supports it. As a result, researchers need ready-to-use miRNA technologies that simplify their research.
Comprehension Obstacles
The general mechanism behind miRNA sounds simple enough: These single-stranded RNAs are complementary to some section of a mRNA, and binding between the two blocks the mRNA from making its protein. As a result, the miRNA reduces the output of that protein.
The details behind the miRNA pathway, though, get more complex. First, an miRNA’s gene and RNA polymerase II produce a string of RNA nucleotides—a primary transcript called pri-miRNA—that includes a double-stranded stem topped with a loop.
An enzyme complex, composed of Drosha and Pasha, cleaves the pri-miRNA to leave only the stem-and-loop section. This so-called pre-miRNA moves from the nucleus to the cytoplasm, where Dicer, another enzyme, clips off the loop. Further processing leaves only a short, single-stranded molecule, miRNA.
Despite unraveling these steps of the biochemical pathway from DNA to miRNA, many details remain unresolved. Beyond the protein complexes that participate in shaping miRNA, other molecules coordinate the cellular translocation from the nucleus to the cytoplasm.
Moreover, the RNA can get edited along the way. In fact, some miRNAs must be edited to be turned on, and others stop working after editing. In addition, mature miRNA might get stored in specific cellular compartments, even going back to the nucleus.
To really understand how miRNAs control gene expression under normal conditions, researchers must refine their knowledge of the mechanism behind this regulation.
For example, scientists want to know the details of how a particular miRNA regulates a specific target, because it might block some targets and merely modulate others. In addition, a better understanding of normal miRNA processes will help scientists understand what causes miRNA regulation to go wrong in a disease and how that problem might be treated or repaired.
In using miRNA as a research tool, though, researchers face further obstacles. For one thing, the short stretch of an miRNA allows it to bind with many mRNAs. In addition, to find out what miRNA does in a specific tissue, a scientist needs a way to get miRNA to the right location. The right spot is not enough, however; it must also avoid the wrong places.
This is particularly crucial in developing miRNA-based therapeutics. For instance, if a scientist developed an miRNA that battles cancer but is also toxic to the liver, it is a useless therapy.
In studying the normal mechanisms of miRNA, researchers must pay attention to tissue-specific functions as well as functions related to a specific developmental stage. Consequently, researchers must note place and time in miRNA-based studies.
Synthetic Solutions
For various forms of miRNA research, scientists need two general tools: mimics and inhibitors. An miRNA mimic can be used to investigate the effect of a specific miRNA—essentially turning on the process that this specific miRNA causes, whether in normal or disease states. On the other hand, an miRNA inhibitor turns off a specific miRNA, which can be used to see what happens when that miRNA is not working.
Recently, Qiagen developed miScript miRNA Mimics and Inhibitors, which are synthetic molecules that are ready for transfection. The mimics are double-stranded RNAs that act like an endogenous miRNA of the same sequence. In other words, these mimics slip into the normal miRNA pathway after the Dicer-catalyzed step, just before the double-stranded miRNA gets turned into single-stranded molecules.
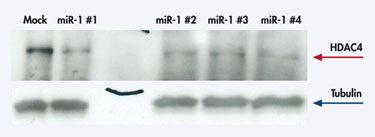
Qiagen scientists have used their miRNA mimics in many experiments such as ones with histone deacetylase 4 (HDAC4), which can modify histones that are involved in the regulation of development and the cell cycle. Transfecting HeLa S3 cells with a mimic for miR-1—an miRNA that targets HDAC4’s mRNA—reduced the levels of this protein (Figure 1).
The miR-1 did not reduce the HDAC4 transcript, however, which indicates that the mimic acts at the translation step (Figure 2).
Qiagen’s miScript miRNA Inhibitors are single-stranded RNAs that block endogenous miRNAs. For instance, HeLa S3 cells were cotransfected with miRNA inhibitors and a reporter construct containing a luciferase gene that was regulated by an endogenously expressed miRNA, and Qiagen scientists found that miScript miRNA Inhibitors impede this endogenous miRNA for at least 96 hours, leading to a regular translation of the luciferase (Figure 3).
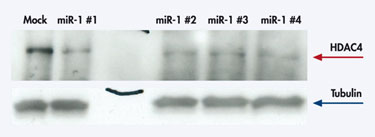
Figure 1 Downregulation of HDAC4 protein after miR-1 mimic transfection.
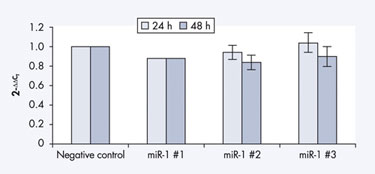
Figure 2 No change in HDAC4 transcript level after miR-1 mimic transfection.
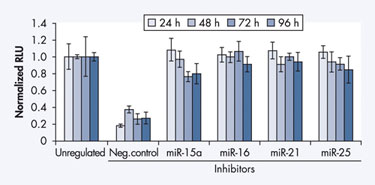
Figure 3 miRNA inhibitor effective after 96 hours.
Mimics and Inhibitors
To deal with the ever-growing list of miRNAs, Qiagen keeps both mimics and inhibitors updated to the current miRBase for human, mouse, and rat. Qiagen also offers custom mimics and inhibitors.
These mimics and inhibitors can be used with a variety of techniques, like detecting and isolating miRNAs. Such standardized tools also simplify functional studies of miRNAs. Moreover, these mimics and inhibitors can enhance downstream applications such as reporter assays, real-time PCR, microarray analysis, and protein analysis.
With real-time PCR, for example, Qiagen used the complete miScript System to quantify miRNA with SYBR Green in HeLa S3 cells. Transfecting these cells with a Qiagen inhibitor dramatically reduced detectable levels of miR-16 miRNA (Figure 4A), and the PCR amplification plot (Figure 4B) also showed the progression of this reduction.
As work on miRNA continues, more applications and tools will be developed. Moreover, the availability of synthetic mimics and inhibitors that work in complete miRNA systems will accelerate this research.
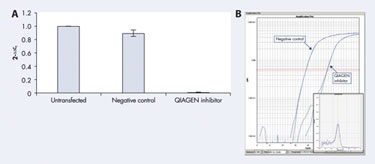
Figure 4A & 4B Decrease in endogenous miRNA after inhibitor transfection.
Constanze Kindler, Ph.D. ([email protected]), is global product manager miRNA at Qiagen. Web: www1.qiagen.com.