June 15, 2009 (Vol. 29, No. 12)
NimbleGen CGH: An Array Portfolio from Discovery to High-Throughput Detection
Comparative genomic hybridization (CGH) was developed to identify pathogenic DNA copy-number changes (e.g., duplications, deletions) on a genome-wide scale, and to map these changes to genomic sequence. Since its first description in 1992, CGH has been rapidly adopted as the method of choice for analysis of DNA copy-number changes associated with cancer, genomic disorders, genetic disease, and other complex phenotypes.
In a typical CGH experiment, genomic DNA isolated from test and reference cell populations are differentially labeled with fluorescent dyes and cohybridized to a target representation of the genome. The relative intensity of test and reference signals at a given genomic position indicates the relative copy number of those sequences in the test and reference genomes.
Initial CGH methods utilized metaphase chromosomes as the target, and the location of DNA copy-number changes between the test and reference genomes were mapped to the physical position on the chromosomes. While this method provided the first efficient approach to scanning entire genomes for DNA copy-number changes, resolution was limited to Mb-sized events and analysis required a high level of cytogenetic expertise.
More recently, high-resolution DNA microarrays have replaced metaphase chromosomes as the target and enabled detection of DNA copy-number changes down to sub-Kb resolution.
With up to 2.1 million (2.1M) oligonucleotide probes per microarray slide, multiplex array formats (e.g., 3 or 12 arrays on a single microarray slide), and flexible array content, Roche NimbleGen CGH arrays enable genome-wide detection of DNA copy-number changes down to ~5 Kb in size and cost-effective analysis of targeted regions at exon-level resolution.
The NimbleGen array CGH workflow is a cost-effective and easy-to-use solution requiring minimal investment in equipment and no prior microarray experience. It can also be easily introduced into most research labs and core facilities. In addition, Roche NimbleGen provides a complete line of instrumentation, reagent kits, and software specifically designed and validated for use with NimbleGen CGH high-density arrays.
Since the beginning, the basic CGH workflow has not changed significantly and includes sample preparation, labeling, hybridization, array washing and scanning, and data analysis and interpretation (Figure 1).
- Sample preparation—Genomic DNA (gDNA) from test and reference cell populations are isolated using standard molecular biology techniques. Unlike other commercially available CGH platforms, NimbleGen CGH arrays utilize full-complexity genomic DNA and do not require DNA complexity reduction, which can introduce bias.
- Labeling—Purified, unamplified, and unfragmented test and reference gDNA are independently labeled with Cy3 and Cy5 fluorescent dyes using a NimbleGen Dual-Color DNA Labeling Kit. The kit is based on a random priming protocol and uses a high concentration of exo-Klenow, lacking 3´ 5´ exonuclease activity, to incorporate dNTPs.
- Hybridization—Test and reference gDNA are cohybridized to a NimbleGen CGH array using a NimbleGen Hybridization System and NimbleGen Hybridization Kit. For multiplex array formats (e.g., 3x720K, 12x135K), Roche NimbleGen offers a Sample Tracking Control Kit that includes control DNAs that are added to the hybridization mix and bind to specific control features on the array. The Sample Tracking controls are used to confirm the sample identity on each array and to ensure experimental integrity.
- Array washing and scanning—NimbleGen CGH arrays are washed using a NimbleGen Wash Kit and scanned at 532 nm (Cy3 channel) and 635 nm (Cy5 channel) using the NimbleGen MS 200 Microarray Scanner. The MS 200 offers scanning down to 2 µm resolution and advanced features such as a 48-slide autoloader, integrated barcode reader, and advanced dynamic autofocus and auto gain for confident and consistent high-quality scans.
- Data analysis—Extraction of raw data from the array image, normalization, and calculation of the log2 ratio of Cy3/Cy5 signal intensities are performed using NimbleScan software. Copy-number gains and losses are identified using the segMNT algorithm, which was developed by Roche NimbleGen and included in NimbleScan software.
- Data interpretation—NimbleGen CGH data are output in a variety of formats including TXT, PDF, and GFF. SignalMap software from Roche NimbleGen provides a simple and interactive data browsing tool to view and interpret GFF files. In addition, data generated from NimbleGen CGH arrays is compatible with a number of software tools available from other vendors (e.g., Nexus Copy Number software from Biodiscovery).

Figure 1. NimbleGen Array CGH workflow featuring the NimbleGen MS 200 Microarray Scanner
Whole-Genome View
NimbleGen CGH arrays are used to identify DNA copy-number changes associated with a broad range of human diseases including mental retardation, autism, schizophrenia, cancer, and autoimmune disease. Figure 2 shows whole-genome PDF views of pathogenic chromosomal gains and losses associated with mental retardation using the NimbleGen CGH 12x135K Whole-Genome Tiling v3.0 Array.
Data is displayed as whole-genome “rainbow” plots where each chromosome is denoted by a different color. CGH analysis of gDNA from a healthy male versus healthy female shows the expected copy-number loss of chromosome X (probes are shifted below the baseline) and gain of chromosome Y (probes are shifted above the baseline), but no other pathogenic copy-number changes are detected. In contrast, large copy-number gains and losses associated with Trisomy 21 and Velocardiofacial Syndrome are detected, as indicated by the arrows.
In addition to whole-genome PDF views, NimbleGen CGH data is provided in GFF for interactive viewing using SignalMap software. Figure 3 shows a zoom-in view of an ~382 Kb deletion region and an ~35 Kb amplification in a Burkitt Lymphoma cell line detected using the NimbleGen Human CGH 3x720K Whole-Genome Tiling v3.0 Array but missed using a lower-resolution competitor’s array.
Data is displayed in SignalMap software alongside annotation tracks (provided with Roche NimbleGen CGH arrays) showing corresponding normal CNVs annotated in the Database of Genomic Variants, segmental duplication regions, and known genes.
The NimbleGen CGH array portfolio includes catalog whole-genome and custom targeted designs in 2.1M, 3x720K, and 12x135K array formats for human and nonhuman organisms (e.g., mouse, rat, dog, cow). This flexibility in array format and design, combined with a simple workflow with validated instruments, reagent kits, and software offers a complete solution for DNA copy-number analysis from high-resolution discovery to high-throughput detection.
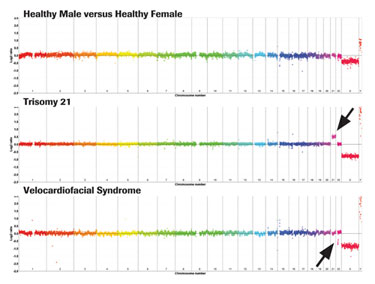
Figure 2. Whole-genome view of NimbleGen CGH data
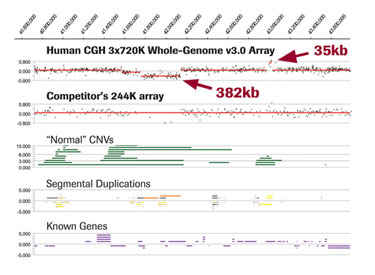
Figure 3. Zoom-in view of NimbleGen CGH data
Vanessa L. Ott, Ph.D. ([email protected]), is CGH product manager at Roche NimbleGen. Web: www.nimblegen.com. Products profiled in this article are for life science research only. NimbleGen is a
trademark of Roche.



