March 1, 2011 (Vol. 31, No. 5)
Advances in Extractin and Enzymology Open Door for RNA as Alternative to Genomic DNA
The Tissue Typing Laboratory tests for compatibility between patients who require an organ or bone marrow transplant and potential donors. In the case of bone marrow transplantation it is important that the tissue type of patient and donor are matched to minimize graft versus host disease. In organ transplantation matching patient and donor tissue types reduces the risk of chronic rejection caused by the development of donor-specific antibodies in the patient against tissue-type mismatches.
A person’s tissue type comprises a set of highly polymorphic proteins called human leukocyte antigens (HLA), which are found on the surface of most cells. HLA plays a central role in adaptive immunity, generating an immune response to pathogens but simultaneously presenting a barrier to marrow and organ transplantation. PCR-based DNA methods have largely replaced serology in tissue typing and, like most laboratories testing large numbers of clinical samples, automation is becoming the norm. In 2006 the New Zealand Tissue Typing Laboratory started using Roche’s MagNA Pure Compact robot to automate DNA extraction.
DNA sequencing has revealed potentially important polymorphisms spread over several exons which, in genomic DNA, can be separated by several kilobases. Tissue typing is becoming an increasingly time-consuming and expensive process using genomic DNA.
We decided to investigate the use of the MagNA Pure robot to isolate total RNA from blood, synthesize cDNA using a Transcriptor cDNA Synthesis kit, and for use as a template for HLA sequencing. RNA is a simpler template than genomic DNA because there are no introns. A random selection of blood samples from transplant patients and donors was selected; these had already been HLA typed using genomic DNA to allow a comparison.
Materials and Methods
Samples and extractions
RNA was extracted from 64 random uncoagulated citrate phosphate dextrose (CPD) blood samples using the MagNA Pure Compact RNA Isolation Kit. The samples were from transplant patients and healthy donors and the blood had been collected from 8 hours prior to extraction, and up to 14 days prior to extraction. Blood samples not extracted immediately were stored at 4°C. Prior to extraction blood samples were thoroughly mixed by several inversions. RNA was quantified using a Nanodrop spectrophotometer.
cDNA Synthesis
In all cases cDNA was synthesized from 0.1 µg (10 µL) RNA using the Transcriptor cDNA Synthesis Kit with random hexamers and oligodT primer.
HLA Amplification and HLA typing
HLA genes, HLA-A, -B, -DQB1, and DRB1, were PCR amplified with AmpliTaq Gold using locus-specific exon primers in exons 1 and 5 (HLA-A and -B) or exons 1 and 4 (HLA-DQB1 and -DRB1).
Nucleotide sequences of amplified exons were determined by cycle sequencing (BigDye Terminator chemistry). Sequenced products were separated by capillary electrophoresis using a 3130XL Genetic Analyzer. Nucleotide sequences were compiled and HLA types assigned using SBTengine, dedicated software that compares heterozygous nucleotide sequences with a library of HLA alleles.
Results
Yield of RNA from 200 µL blood was the same (0.1–1 µg approx) regardless of age of blood. The HLA types obtained by sequencing were concordant with those obtained by HLA typing genomic DNA using Luminex. A small number of examples are shown in the Table with the HLA-A, -B, -DR, and -DQ types of three samples. No preferential amplification of alleles, or allele drop-out, was observed. The ambiguities observed are identical to those observed in genomic DNA SBT, as expected.
Ambiguous SBT results are often seen when a “common” allele group is present where the heterozygous sequence of one pair of alleles is identical to at least one other pair of alleles (for HLA-A, -DRB1). In the case of HLA-B the ambiguity arises due to an additional polymorphism found in exon 4, which was not sequenced in the protocol described in this article.
It is debatable whether polymorphisms outside of the peptide binding domains encoded by exon 2 (in HLA-DQB1 and -DRB1) or exons 2 + 3 (HLA-A, -B) are clinically relevant. It is acceptable to report the pair of alleles most common in your population and record the others as “cannot exclude”. Routine HLA-A,B,DR typing by Luminex generally produces a medium resolution result with a number of possible HLA alleles that are abbreviated into a letter code (Table). HLA-DQB1 was typed by PCR-SSP of genomic DNA.
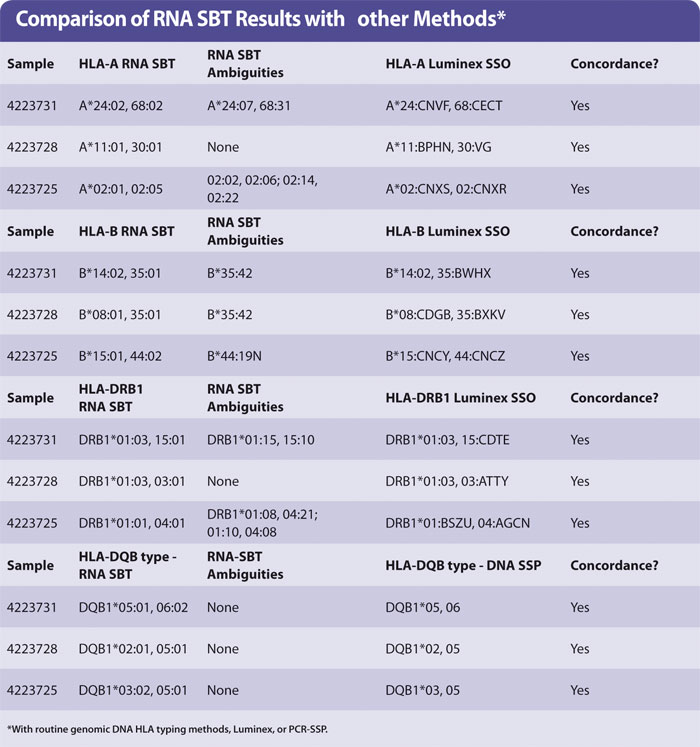
Figure 1 shows a sequencing electropherogram (from SBTengine) covering a short sequence in exon 2 of HLA-DRB1. In this sample exons 1, 2, and 3 were easily sequenced using RNA as template. In genomic DNA this would be quite a feat as exons 1 and 3 in HLA-DRB1 are between 6 and 10 kbp apart, depending on the alleles. In the example shown this allowed resolution of DRB1*14:01/14:54 to DRB1*14:54 and these two alleles only differ in exon 3. To resolve these two alleles one would have to purchase a high-resolution DR14 SSP kit or employ exon 3 amplification prio to sequencing of genomic DNA.
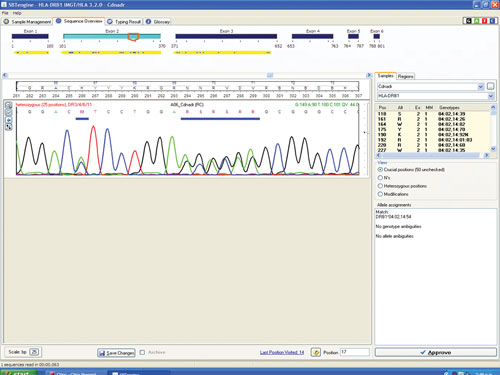
Figure 1. SBTengine sequence chromatogram of HLA-DRB1 exon 2
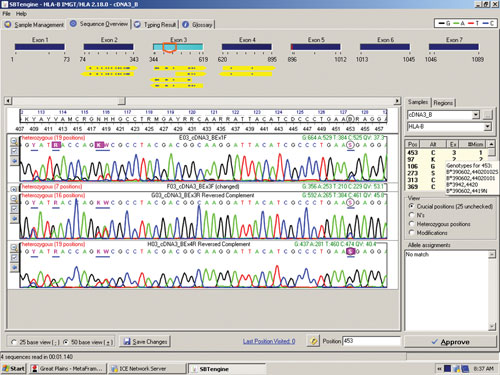
The sequence in Figure 1 clearly shows several heterozygous sequence positions. Among the 64 samples we identified one new HLA-B allele, B*44:59, which had not been identified when this sample was previously HLA typed using genomic DNA. This unique heterozygous position defining B*44:59 (in combination with the other allele) is shown in Figure 2.
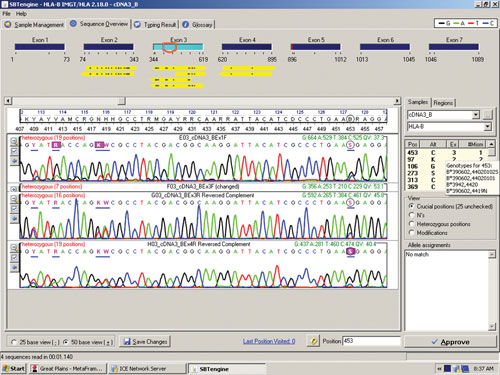
Figure 2. RNA sequencing identifies a new HLA-B allele, B*44:59
Conclusions
Advances in extraction technology and enzymology mean that RNA now offers a realistic and simpler alternative to genomic DNA in the Tissue Typing Laboratory. The RNA method will also be more cost effective. The method can be simplified further by including cDNA synthesis and PCR in one tube. The method should also be amenable to automation after RNA extraction.
HLA genes are readily amplified from cDNA, sequenced, and typed from the cDNA (most are heterozygous). The tissue types are in agreement with the DNA-based types for these samples. With no introns this method offers a much simpler sequencing strategy with only 2–4 sequencing reactions compared to 8–12 when genomic DNA is used as template.
Using RNA as a template for HLA typing is also a useful tool for checking for the expression of HLA alleles, which may be missed using genomic DNA. We have used this approach to characterize a new null allele, HLA-A*01:34N, which has a novel splice site in exon 4. Genomic DNA sequencing showed only a silent substitution in exon 4 (GCG instead of “normal” GCA) but sequencing cDNA showed that exon 4 transcribed from this allele started after this SNP, as it had created an alternative splice site.
Marrean Theseira is medical laboratory scientist and Paul Dunn ([email protected]) is clinical scientist at the Tissue Typing Laboratory at the New Zealand Blood Service.