February 1, 2011 (Vol. 31, No. 3)
New Platform Designed to Accelerate Subunit Vaccine Development for Infectious Diseases
As we are continually confronted by the devastating effects of emerging and re-emerging diseases such as malaria, multidrug-resistant tuberculosis, and HIV, research and development to find effective vaccines is becoming more important than ever.
Recombinant proteins in combination with novel adjuvants, so-called subunit vaccines, have become a major focus. Creation of these vaccines depends on the identification of appropriate subunit antigens (or epitopes) that the immune system can effectively target. Genomic and proteomic-based technologies are used to identify a number of antigen candidates, from which immunogenic, well-defined antigens can then be chosen for further development.
The criteria to select such antigens depends on certain characteristics such as pathogen conservation, human immunogenicity without compromising safety, and, in many cases, induction of functional antibodies.
Methods to rapidly and efficiently evaluate protein antigenicity are central to the identification of appropriate targets from a large number of antigen candidates, especially regarding the extent to which these candidates can be recognized by the adaptive immune system through T- and B-cell responses.
To address this requirement, ProImmune has developed a rapid antigen-characterization platform that encompasses B-cell epitope prediction and B-cell linear epitope mapping service modules, both of which result in a relative ranking of antigenic regions within proteins or between different proteins in a group.
The T-cell epitope identification and validation process employs MHC-peptide binding and rate assays, antigen-specific T-cell detection by flow cytometry in both preclinical animal models and human samples, and functional cellular assay services such as ELISpot and intracellular cytokine staining. Additionally, assessment of the relative antigenicity of candidate subunit vaccines is enabled by naïve T cell and dendritic cell (DC) -T cell proliferation assays.
Defining and Ranking B-Cell Epitopes
ProImmune’s B-cell epitope prediction system is based on primary protein sequence and, where available, three-dimensional structure data. It uses an intelligent consensus approach that employs a collection of up-to-date public domain and commercially licensed scientific databases and software. Importantly, this method can also be used to compare different clinical strains of the organism, to discover whether the putative epitopes are conserved.
The system also allows screening of overlapping peptides generated from the pathogen protein sequences of interest, against samples of serum from patients with a well-characterized response. These results can then define and rank B-cell linear epitopes. This approach identifies antigens that are expressed during the course of the disease, a prerequisite of a subunit vaccine for conferring protection.
T-Cell Epitopes
In the human immune system, different cells are responsible for specific types of immune responses. Cytotoxic (CD8+) T-cell responses kill infected target cells, whereas helper (CD4+) T cells provide help to memory B cells producing antibodies. Those cells mount their immune responses by recognizing MHC-epitope complexes on the surface of target cells. The discovery and characterization of these T-cell epitopes is challenging, and the limitations of traditional methods that rely on functional cellular assays make projects labor intensive and time consuming.
To address these issues, ProImmune has developed technologies that allow screening of any protein for T-cell epitopes in weeks, instead of months or years. The technology combines cell-free in vitro Reveal™ MHC-peptide binding and rate assays with the synthesis of ProVE® MHC Class I Pentamers for conclusive confirmation of epitopes on vaccine clinical cell samples. Class II epitopes can be validated using a suite of comprehensive functional assays.
The approach determines the exact binding sequence and MHC restriction of the potential epitope in a single step and the likelihood of a peptide being presented long enough for it to represent a good T-cell epitope. This pivotal information can be combined with binding assay data to identify which peptide-epitopes will be important in the vaccine development strategy. Furthermore, if cell samples are in limited supply for final epitope validation, rate assay data can be used to prioritize candidates, resulting in the use of smaller sample volumes. This enables subunit vaccine discovery projects to progress rapidly and economically.
Identification in HIV-1
In HIV immunity, factors that potentially distinguish between protective and ineffective anti-HIV-1 immune responses and control disease progression relate to epitope-MHC binding as well as stability and affinity. Researchers at ProImmune recently supported a study carried out by Samantha Westrop and her colleagues at Imperial College to examine the details of HIV-1 CD8+ T-cell epitope recognition. This study used rapid peptide synthesis, Reveal binding and affinity assays, and ProVE Pentamer synthesis in conjunction with flow cytometry, as well as ELISpot analyses to discover new T-cell epitopes for HIV-1.
The rate of HIV-1 disease progression is also influenced by the genetic profile of an individual. For example, individuals with HLA-B35 and HLA-B8 profiles have been associated with an increased rate of disease progression in HIV-1+ infections. To discover possible reasons for this rapid disease progression, ProImmune’s methods were used to investigate the HIV-1 Gag peptide-specific CD8+ T-cell immune response mounted by HLA-B35+ individuals. Initially, forty-four 9-mer HIV-1 Gag peptides were synthesized as a PEPscreen® Custom Peptide Library, and were assessed for binding to B*35:01 using the Reveal MHC-peptide binding assay.
The results implicated 12 peptides as potential epitopes (Figure 1), and these were further analyzed with the Reveal rate assay to determine their rates of dissociation from the MHC complex. Two of the 9-mer peptides, YPLTSLRSL (peptide 1) and HPVHAGPIA (peptide 8) were found to form a stable complex with B*35:01, indicating that these may be potential novel epitopes.
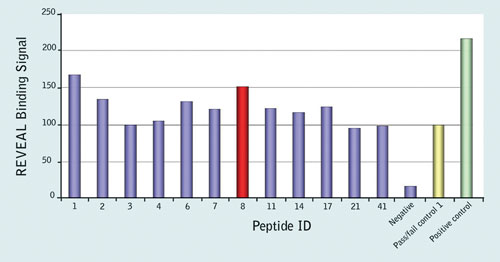
Figure 1. Results of the Reveal MHC-peptide binding assay for controls and the 12 peptides that were considered potential epitopes; binding signal is shown as a percentage relative to the pass/fail control; peptide 8, HPVHAGPIA (red) was further validated by MHC Class I Pentamer staining.
Pro5® MHC Class I Pentamer staining was then carried out on patient samples, which confirmed the presence of a well-defined Pentamer+/CD8+ population for the B*35:01/HPVHAGPIA epitope (Figure 2). Additionally, no naïve or central memory T cells specific for the HPVHAGPIA peptide were found, indicating a lack of proliferative potential in infected HIV-1+ individuals. Further validation with ELISpot assays showed that the HPVHAGPIA peptide elicited the highest ex vivo response of the peptides studied.
Additionally, using the Reveal and ProVE system, a previously unknown epitope was identified, which may be involved in an immune response in B*35:01-positive individuals. The results of this study advance our understanding of HIV-1 immunopathogenesis and disease progression and contribute toward the ultimate goal of vaccine development.
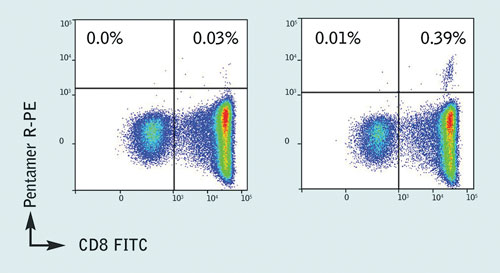
Figure 2. Pro5 Pentamer staining of live lymphocytes gated on CD3+ cells; the left plot shows staining with an allele mismatched negative control Pentamer, the right plot shows staining with the HA9 (B*35:01/HPVHAGPIA) Pentamer. 0.39% of CD3+ live lymphocytes are CD8+/ B*35:01/HPVHAGPIA Pentamer+ with a background stain of 0.03%.
Summary
ProImmune’s rapid antigen-characterization platform encompasses complementary B- and T-cell epitope discovery systems and functional cellular assays such as T-cell proliferation and ELISpot. Not only do these technologies characterize antigenicity of proteins in terms of T- and B-cell responses and allow ranking and selection as part of the overall epitope identification effort, but they also establish the key biomarkers for measuring responses in downstream clinical work.
Amanda Turner ([email protected]) is marketing operations manager, and Nikolai Schwabe, Ph.D. ([email protected]), is CEO at ProImmune.



