September 15, 2013 (Vol. 33, No. 16)
Novel Instrument Automates Critical Procedure Using Chemical Denaturation
The requirement to formulate biologics in a safe and stable form, and the known propensity for these molecules to aggregate, has led to the need for rigorous stability characterization of these biotherapeutics. This characterization is conducted using a wide range of bioanalytical and biophysical techniques. Combined, these techniques aim to determine the protein stability profile as well as the propensity for aggregation.
Since protein denaturation often leads to aggregation, improving structural stability is important to retard or prevent the formation of aggregates.
There are multiple ways to measure the structural stability of proteins, and they all involve disrupting the protein structure by either physical or chemical means. Temperature is one of the most widely used physical denaturation tools; its main drawback is that proteins usually denature at 60ºC or higher temperatures while the temperature of interest is either physiological temperature (37ºC) or a storage temperature (4ºC or 25ºC), requiring result extrapolations of 20–55º. Temperature extrapolations are error prone because the temperature stability of proteins is a function of three parameters: enthalpy, entropy, and heat-capacity changes.
Heat-capacity changes, in particular, are difficult to measure. The preferred method, differential scanning calorimetry—is not accurate at low protein concentrations. In addition, temperature denaturation is often irreversible and accompanied by aggregation and precipitation, precluding a rigorous thermodynamic analysis. Under conditions of irreversibility—generally the case with antibodies—it is impossible to extrapolate reliable stability parameters to lower temperatures. Consequently, stability rankings have been traditionally presented in terms of Tm values.
As illustrated in Figure 1, providing a thorough evaluation of protein stability requires confirmation of thermal analysis results using an independent and completely orthogonal method.
Here we will discuss a different approach, chemical denaturation, as a tool in the optimization of formulation conditions for biologics, and how chemical denaturation complements the role of thermal denaturation for this purpose. Chemical denaturation has been proven to provide reliable thermodynamic values for protein stability for over 40 years.
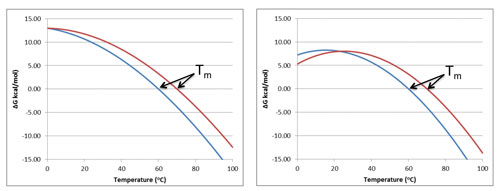
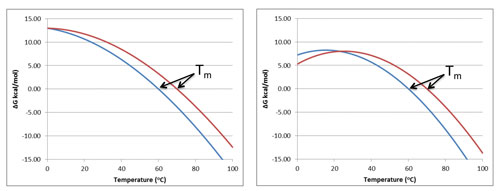
Figure 1. The temperature dependence of the Gibbs energy of protein stability calculated for typical protein parameters: The curvature of the temperature dependence depends on the heat capacity change ΔCp. The left panel illustrates the case for two proteins both with a ΔCp of 1.5 kcal/K*mol. In this case the observed 10°C difference in Tm is indicative of the relative protein stabilities at all temperatures down to 0ºC. The right panel illustrates the case for two proteins both with a ΔCp of 2.5 kcal/K*mol. A larger ΔCp is indicative of a higher degree of hydrophobic exposure upon unfolding. Although the same 10°C difference in Tm is observed, there is an actual reversal of the stability rank order at room temperature and below. Accurate determination of ΔCp requires higher protein concentrations. In addition, for irreversible thermal denaturation, the actual ΔCp value is unknown. In all cases ΔH at Tm was assumed to be 120 kcal/mol at 60°C and 70°C, respectively. This example illustrates the need to confirm thermally observed stability rank-order determinations with an independent, orthogonal technique.
Automated Stability Determination
A dedicated instrument, the Model 2304 Automated Protein Denaturation System, which completely automates stability determination using chemical denaturation, has been developed by AVIA Biosystems. This system uses intrinsic (or extrinsic) fluorescence to monitor the protein conformational change associated with protein unfolding (denaturation).
As illustrated in Figure 2, protein stability curves are generated by monitoring fluorescence as a function of increasing denaturant concentration.
Identifying the solvent conditions that maximize the structural stability of biologics can be completely automated on the 2304 using the chemical denaturation technique. Solution conditions such as buffer composition, buffer strength, pH, ionic strength, excipient composition, excipient concentration, and protein concentration can be evaluated for their effect on protein stability. For formulations and process-development applications, up to 96 different sets of conditions can be evaluated for stability in a single automated run.
The actual formulations or process condition solutions being tested may be prepared manually and placed on the instrument, or they can be automatically prepared by the instrument from stock solutions. It is also possible to evaluate up to 96 different protein constructs for relative stability. In each case, all the sample preparation, data collection, data analysis, and stability report generation is automatically performed by the instrument.
Figure 2. Protein stability curves are generated by monitoring fluorescence as a function of increasing denaturant concentration. Formulation conditions that shift the stability curve to the right (toward higher denaturant concentration) indicate an increase in protein stability. Conversely, formulation conditions that shift the stability curve to the left indicate a decrease in protein stability.
As illustrated in Figure 3, fluorescence data is collected at a series of denaturant concentrations, and the raw data is fit to a protein unfolding transition state model. The calculated thermodynamic parameters ΔGº, m, and C1/2 are determined by nonlinear least-squares fitting of the data to the model. ΔGº is the Gibbs free energy of protein unfolding in the selected buffer, m is the rate of change in ΔG as a function of denaturant concentration, and C1/2 is the denaturant concentration at which one-half the protein molecules are in the native state and one-half are in the unfolded state. Typically, ΔGº and C1/2 increase as the stability of a biologic is improved. Protein constructs, formulations, and process conditions can all be rank-ordered for stability using the ΔGº and C1/2 parameters.
Chemical denaturation can also be measured with other physical observables like ultraviolet spectroscopy, dynamic light scattering, or circular dichroism. In addition to ΔGº, m, and C1/2 values, these different measurements can provide specific structural information.
For example, while tryptophan fluorescence provides information about the solvent exposure of those residues, circular dichroism reports changes in secondary structure. Different endpoint values may be used to assess the degree of unfolding or the environment of the fluorophores upon denaturation. In the case of tryptophan fluorescence, this additional information is contained in the emission spectrum.
Fluorescence measurements can also be performed with extrinsic fluorescence probes (such as ANS, Sypro Orange, Nile Red, and Thioflavin T), which have the added advantage of being sensitive to additional processes such as aggregation or the formation of protein fibers. Whether using either intrinsic or extrinsic fluorescence, the new instrument completely automates the chemical denaturation experiment. When used in conjunction with other instruments (such as dynamic light scattering or circular dichroism), the 2304 can be used for automated sample processing for subsequent off-line analysis.
In addition to rank-ordering by stability, biologics can be rank-ordered by propensity to aggregate. This application is also accomplished automatically by the AVIA instrument. In this case, the denaturant (typically urea or guanidinium chloride) is substituted with an ammonium sulfate solution. By monitoring 90º light scattering (using the same detector) as a function of ammonium sulfate concentration, different protein constructs or different formulations can be rank-ordered directly by propensity to aggregate.
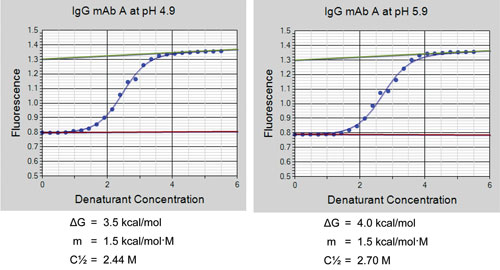
Figure 3. Denaturation curves for an IgG mAb at pH 4.9 (left panel) and pH 5.9 (right panel). Displayed curves contain 24 raw fluorescence data points (blue circles), which are automatically fitted to a transition state model (blue line) using a nonlinear least-squares fitting method. The red lines are the native protein base lines, and the green lines are the unfolded protein base lines. The thermodynamic parameters ΔG, m, and C1/2 are automatically calculated from the fitting of the data to the transition state model.
In pharmaceutical discovery and development settings, large numbers of constructs or formulations need to be rapidly and easily compared (rank-ordered) for stability. To facilitate this, fraction denatured curves are automatically generated for all samples. As illustrated in Figure 4, fraction denatured curves can be selected and overlaid to provide a direct visual comparison of the effect on protein stability of different changes to the protein construct, the formulation, or the process conditions.
The advent of instrumentation with the capability of automatically performing sample preparation, measurements, and data analysis makes the well-established chemical denaturation technique viable and practical in virtually any laboratory setting. There is now a practicable means to complement thermal screening results with a fully orthogonal method and measurement system for protein stability determinations.
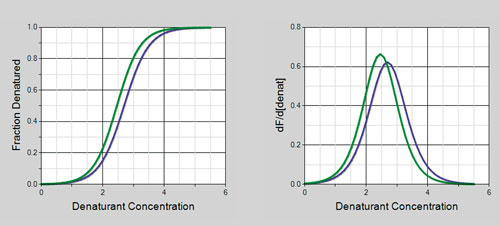
Figure 4. Fraction denatured curves for the IgG mAb at pH 5.9 (blue lines) and pH 4.9 (green lines): These curves are automatically generated to facilitate the stability comparison of multiple samples. The left panel is the fraction denatured curve and the right panel is the first derivative of the fraction denatured curve. The sample at pH 5.9 is more stable (that is, has a higher C1/2 value) than the sample at pH 4.9.
Richard Brown ([email protected]), president, Ernesto Freire, CSO and Henry Walters professor of biology at The Johns Hopkins University, and Burleigh Hutchins, CTO, co-founded Avia Biosystems.