March 1, 2008 (Vol. 28, No. 5)
Vast Increases in Speed and Versatility Are on the Horizon
When Leonard Bloksberg, Ph.D., was a research biologist in New Zealand, his large genomic searches using a DEC Alpha Cluster and BLAST were estimated to take years. As Dr. Bloksberg observed during Cambridge Healthtech’s“Next-Generation Sequencing” (NGS) conference, held late last year in Providence, RI, the genomics-data monster doubles every six months. Computational power, following Moore’s law, however, doubles every 18 months, and BLAST is a 16-year-old technology.
These facts motivated Dr. Bloksberg to begin development on what was to become the SLIM Search™ technology. Working with a group of computer scientists and mathematicians, search time was soon reduced from years to three hours. Encouraged by this, Dr. Bloksberg decided to start the company SLIM Search to provide the biotech industry with fast pattern-matching and genomic-search solutions.
SLIM Search, he told conference attendees, “can provide up to 10,000 times more speed than BLAST. It returns richer data content, which makes it possible to create scores and molecular alignments directly from a word search,” he said. SLIM Search also tags the alignments digitally, so subsequent analysis can be computerized and automated for the first time.”
Quicker Sequence Similarity Searching
Until a few years ago, almost all sequencing was done using Applied Biosystems’ instruments, noted Brian Hilbush, technical director of SLIM Search’s U.S. subsidiary. Celera, for example, “had 500 ABI 3700 instruments to sequence the human genome,” said Hilbush. Applied Bio’s instruments, along with those of several other leading players, continue to be key next-generation sequencing technology.
BLAST and SLIM Search are distinct algorithms and utilize different approaches to sequence similarity searching, Hilbush explained. BLAST preprocesses the query and invokes a dynamic programming matrix for creating local alignments. SLIM Search, on the other hand, builds highly stable arrays on the subject database and reports alignments in a novel format, he explained.
A fundamental difference, Hilbush pointed out, is that BLAST rebuilds for each run, while SLIM Search does one build and then scans the database with all data stored in RAM. BLAST produces the widely recognizable standard ASCII graphical output for pair-wise alignments.
SLIM Search, he noted, is compatible with all NGS platforms. Sensitivity filters allow searches for short, 30–50 base pair sequences, and can be optimized for mapping and anchoring these reads to complex genomic reference sequences. The software incorporates special features suited for comparative genomics, genome assemblies, orthogonal gene mapping, phylogenetics, and SNP analysis.
Droplet-Based Microfluidics Platform
Talking about technology involved in one of the initial steps in NGS, sample preparation and amplification, John H. Leamon, Ph.D., project lead, nucleic acid applications at RainDance Technologies (RDT), discussed the firm’s new fluid-handling technology. The product is based on the use of picoliter-sized droplets to encapsulate samples for a wide range of molecular assays.
RDT will commercialize the droplet-based microfluidics platform as the professional laboratory system (PLS) brand. The company expects to generate revenue in 2008 from sales of a genome-selection instrument and custom primer libraries in a droplet format that will provide rapid, high-throughput sample amplification with equimolar amplicon concentrations for targeted sequencing using a variety of next-generation sequencing systems.
Dozens of current generic and specialized genomic, drug discovery, diagnostic, and industrial applications can be ported over to RDT’s microdroplet format, Dr. Leamon noted. The company’s microfluidic chips can be used to replace a range of fluid-handling and analytical-chemistry instruments. RDT also plans to exploit its platform’s ability to detect very low copy-number biomarkers in cancer cells by amplifying the assay signal within each sample droplet.
When the PLS is configured for a specific application, it can provide reagent input/output connections, a detector system with up to three lasers, a graphical user interface, and a chip dock into which molded silicon chips are inserted for processing and analysis. Application-specific chips and instrument configurations provide system versatility. For sequencing prep applications, for example, the PLS can do isothermal amplification, solid-phase PCR, solution-phase PCR, and multiplex PCR.
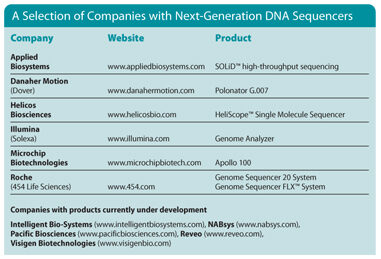
Next-Generation DNA Sequencers
The nanoreactor droplets, comprising aqueous samples, flow through the chips in a stream of immiscible oil containing surfactant stabilizers. There is no air or chip interface, and the droplet samples are stable when stored at room temperature or subjected to PCR amplification. Droplet volume range is 0.5 picoliters to 100.0 nanoliters, with droplet size ranging from 5–500 µm in diameter. Throughput is 1,000–10,000 droplets per second.
A wide range of microfluidic elements enable the system to perform various functions common to most assays such as generating droplets; formulating droplets with multiple reagents; as well as combining, mixing, incubating, detecting, storing, and thermally cycling the stream of droplets.
For targeted-sequencing sample prep, droplet primer libraries combine individual primer pairs with DNA sample droplets to enable de facto multiplex PCR for thousands of amplicons. “You pull the DNA you need for PCR and combine it with primer,” Dr. Leamon explained. “Primers merge with DNA droplets in an electric field, after which you do your PCR amplification. Our initial work used two-step PCR with 95º and 68ºC temperature zones on the chip. With 100 picoliter droplets, thermal transition times are rapid, and the PLS can process 3–10 million primer specific PCR reactions per hour.”
De Novo Genome Sequencing
Gabor Marth, D.Sc., assistant professor of biology at Boston College, foresees NGS applications in genome resequencing like in somatic-mutation detection, organismal-SNP discovery, mutational profiling, structural-variation discovery, as well as de novo genome sequencing. Short-read sequencing will at least be an alternative to microarrays for DNA-protein interaction analysis, novel transcript discovery, quantification of gene expression, and epigenetic analysis using methylation profiling, he added.
Andreas Sundquist, a doctoral student under Serafim Batzoglou, Ph.D., assistant professor in the computer science department of Stanford University, reiterated that distinct sequencing strategies are required for resequencing where the objective is to understand how one individual in a species differs from the others and de novo sequencing on genomes that have never been sequenced before, for which mammalian sequencing is still costly and time consuming.
Dr. Batzoglou’s group uses 454 Life Sciences sequencing technology for short reads and faster, more cost-effective results. SHRAP, or short read assembly protocol, uses a random library of clones and sequences them. Reads are identified with the clones from which they came. The genome is covered by clones to 10-fold coverage, and each clone is sequenced by reads to twofold coverage. From the read data alone, it is possible to figure out how the clones overlap and assembly can proceed in larger regions.
As Sundquist’s presentation revealed, coverage is significantly more important than read length, with all metrics favoring 20X coverage at 200 bp read length over 11.25X coverage at 250–300 bp. Sundquist summarized by stating that Stanford’s protocol is suitable for high-throughput automation and that it is possible to assemble a de novo repetitive mammalian genome with such reads.
Data-Management Challenges
Likening the challenge to solving a large jigsaw puzzle that is all sky pieces, Sundquist presented a novel protocol for whole-genome sequencing. Since all genomes are limited to four nucleic acids, sequences in complex genomes are highly repetitive. In the department of computer science, the group led by Dr. Batzoglou used a scalable variant on hierarchical sequencing to solve repetitiveness, Sundquist explained.
Taking up data-management issues, Dr. Marth commented, “just determining how to index data on a disk so you can access it serially is a significant task.” Interpreting machine readouts such as base calling and base error estimation are among the fundamental informatics challenges NGS researchers face.
Various platforms are more liable to make insertion/deletion (INDEL) errors, while others are more prone to substitution errors. Dr. Marth noted that 454 technology features somewhat longer read lengths (20–100 mb in 100–250 bp reads) and therefore lower throughput than both Illumina and Applied Bio (1–4 gb in 25–50 bp reads) and that the former results in somewhat higher INDEL errors but fewer substitutions. In nonunique areas of the genome, he pointed out, it is sometimes impossible to know which area is represented with only 25 to 50 base pairs.
Based on a partnership with Doug Smith, Ph.D, at Agencourt Bioscience, Dr. Marth reported the results of mutational profiling using deep 454/Illumina/SOLiD data with Pichia stipitis. This organism converts xylose to ethanol and is of interest for biofuel production.
The collaborators found that one mutagenized strain had especially high conversion efficiency. To determine where the mutations were that created this phenotype, the group resequenced the 15 mb genome with reads from the three machines and confirmed 14 true point mutations in the entire genome.
Deepak Thakkar, Ph.D., bioscience solutions manager at SGI, described supercomputing solutions developed to address major challenges in areas such as data management, scientific productivity, and data storage. There are three types of SGI solutions: Altix® XE Clusters for high-throughput processes, Altix Shared Memory System for high-performance processes, and RASC™ FPGA Solutions. In addition, SGI InfiniteStorage solutions provide the underpinning to enable fast data access as well as continuous data growth and to scale seamlessly in a mixed computing environment workflow as data requirements increase exponentially.
Depending on the application, any one of the three basic approaches is used. “Our hybrid solution is a different way of looking at scientific workflow,” Dr. Thakkar remarked. Bioinformatics and chemistry applications, for example, utilize different scientific codes. Data input is therefore directed to the cluster that is optimal for a specific application based on computational-platform efficiency. “The system intuitively directs data flow like an intelligent master scheduler,” he commented.
Specifically addressing bioinformatics, Dr. Thakkar said that Altix 450 and RASC field-programmable gate-array technology accelerate small queries of approximately 25 nucleotides 60 times over Opteron clusters and are about 19 times faster for large queries of approximately 115 nucleotides. Additionally, power consumption is claimed to be 90% lower for the SGI system than for Opteron clusters.
Dr. Thakkar also detailed the performance advantages of SGI Altix ICE, a next-generation platform that is billed as “a tightly integrated, cool-running blade solution.” Designed by the company to close the growing gap between performance and user productivity, the new platform is the first in a new line of bladed servers purposely built to handle true high-performance computing applications and large scale-out workloads, he reported.